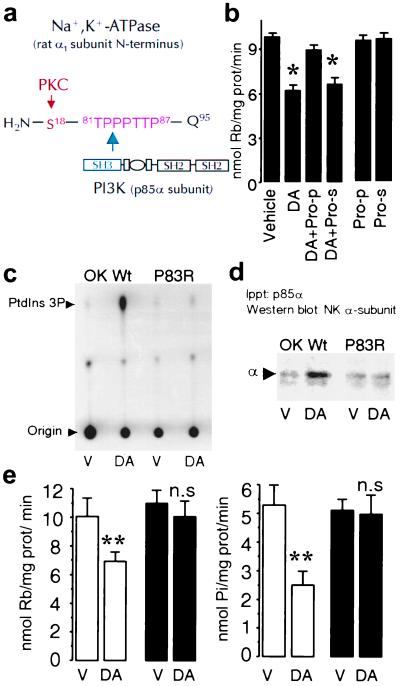
Figure 3.
A proline-rich motif within the Na+,K+-ATPase α1 subunit is necessary for binding and activation of PI3K. (a) Schematic representation of the rat Na+,K+-ATPase α1 subunit N-terminal with the predicted amino acid (Gln95) adjacent to the plasma membrane. A proline-rich motif (TPPPTTP87) is located in the vicinity of the substrate (Ser18) for PKC. (b) Na+,K+-ATPase activity was determined in OK-Wt cells incubated with 1 μM DA in the presence or absence of a peptide (50 μM) comprising the proline motif (Pro-p) or of a scrambled peptide (Pro-s). Each bar represents the mean ±SEM of three experiments performed in triplicate. *, P < 0.02. (c) PI3K activity was determined as described (see legend to Fig. 1) in vehicle (V)- or DA-treated cells. (d) The presence of Na+,K+-ATPase α1 subunits in the immunoprecipitates was determined by Western blotting with a monoclonal antibody against the Na+,K+-ATPase α1 subunit as in Fig. 1. The data are representative of four experiments. (e) Na+,K+-ATPase activity was determined either as Rb+ transport (Left) or ATP hydrolysis (Right). Cells expressing the wild type (open bars) or the P83R mutant (filled bars) were incubated with vehicle (V; Hanks' buffer) or 1 μM DA. Each bar represents the mean ±SEM of five experiments performed in triplicate. **, P < 0.01. n.s., Not significant.