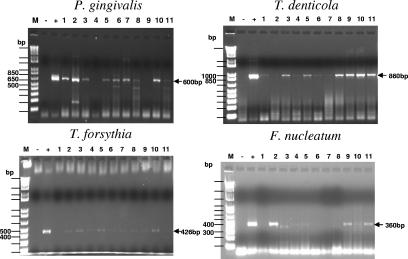
FIG. 2.
PCR analysis of oral microbial samples from rats infected with a polymicrobial inoculum (P. gingivalis, T. denticola, and T. forsythia or P. gingivalis, T. denticola, T. forsythia, and F. nucleatum). Agarose (1.5%) gels contained PCR products from reactions with P. gingivalis, T. denticola, T. forsythia, and F. nucleatum primers. Lane M, 1 Kb Plus DNA ladder; lane −, negative control containing the appropriate bacterial primer but no target DNA; lane +, positive control; lanes 1 to 11, oral microbial samples from individual infected rats. For P. gingivalis 381, a 600-bp amplicon was obtained and 9 of 11 infected rats were positive for P. gingivalis after the third infection. For T. denticola ATCC 35404, a 860-bp amplicon was obtained and all 11 infected rats were positive for T. denticola after the fourth infection (a few bands were faint). For T. forsythia ATCC 43037, a 426-bp amplicon was obtained and 9 of 11 infected rats were positive for T. forsythia after the fourth infection (a few bands were faint). For F. nucleatum ATCC 49256, a 360-bp amplicon was obtained and 7 of 11 infected rats were positive for F. nucleatum after the third infection (a few bands were faint).