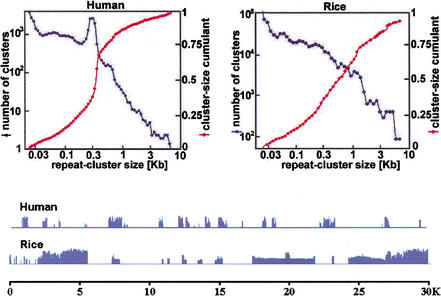
Figure 5.
Repeat-cluster size characteristics. Clusters are defined by placing the 20mer repeats, determined from the shotgun data, onto 11.9 Mb and 0.89 Mb of finished human and rice bacterial artificial chromosome sequence, respectively. Any 20mers separated by <26 bp of unique sequence are merged together, and it is the sizes of these merged clusters that are plotted. In the distribution function for human, the peak near 300 bp is due to Alu transposons. In rice, the distribution is scaled up to reflect the entire rice genome. The cumulants show that a significant fraction of rice repeats lie in kilobase-sized clusters. Another way to demonstrate this fact is to highlight 20mer repeats by blue histogram bars proportional to copy number in typical human and rice segments.