Abstract
The primary membrane of vaccinia virus, as well as those of other poxviruses, forms within a discrete cytoplasmic factory region. We recently determined the existence of an operative pathway from the endoplasmic reticulum within the virus factory to nascent viral membranes and demonstrated that a viral protein could be diverted from this pathway to Golgi membranes by the addition of COPII-binding sites (M. Husain, A. S. Weisberg, and B. Moss, Proc. Natl. Acad. Sci. USA, 103:19506-19511, 2006). Here we describe an investigation of the structural features that are required for transit of proteins to the viral membrane. Deletion of either the N-terminal domain or the C-terminal cytoplasmic tail from the conserved A9 protein did not prevent its incorporation into viral membranes, whereas deletion of the transmembrane domain resulted in its distribution throughout the cytoplasm. Nevertheless, replacement of the A9 transmembrane domain with the corresponding region of a nonpoxvirus transmembrane protein or of a vaccinia virus extracellular envelope protein allowed viral membrane targeting, indicating no requirement for a specific amino acid sequence. Remarkably, the epitope-tagged A9 transmembrane domain alone, as well as a heterologous transmembrane domain lacking a poxvirus sequence, was sufficient for viral membrane association. The data are consistent with a sequence-independent pathway in which transmembrane proteins that are synthesized within the virus factory and lack COPII or other binding sites that enable conventional endoplasmic reticulum exiting are incorporated into nascent viral membranes.
Assembly of vaccinia virus (VACV) and other poxviruses begins with the formation of a crescent-shaped membrane that enlarges into a spherical immature virion (IV), which then condenses into a brick-shaped infectious mature virion (MV) (3, 4, 8). The MV consists of a core, containing the DNA genome as well as RNA polymerase and transcription factors, surrounded by a lipid membrane with more than 20 proteins, none of which are glycosylated. Some MVs that move out of the factory are wrapped with modified trans-Golgi or endosomal cisternae containing glycosylated viral proteins unrelated to those of the MV and then transported to the plasma membrane and liberated as extracellular virions (EVs), which are essentially MVs with an additional outer membrane (27). The MV membrane can fuse with either the plasma membrane or the membranes of endocytic vesicles, whereas the EV membrane is not fusogenic and must be disrupted prior to virus entry (1, 14, 16, 25, 35).
The inability to discern clear continuity of the viral crescent membrane with a cellular organelle (4, 7, 8, 10) led to speculation that the viral membrane is formed by a novel mechanism. Skepticism regarding de novo membrane formation, however, inspired efforts to obtain evidence for the origin of the IV membrane from a component of the cellular secretory pathway. MV transmembrane proteins in the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) and ERGIC proteins in tubules or vesicles near crescent membranes were detected by immunogold labeling (5, 6, 13, 20, 22, 28-30), although trafficking of proteins between cellular and viral membranes was not demonstrated. The finding that expression of a dominant-negative form of Sar1 GTPase, which arrests COPII transport, had no effect on IV and MV production suggested that the viral membranes could arise directly from the ER (11). However, the absence of signal peptide cleavage or glycosylation made it difficult to confirm either ER or ERGIC trafficking by biochemical methods. To address this problem, we constructed a chimeric protein in which the N terminus of the A9 MV protein was replaced with a heterologous signal peptide; the latter was cleaved, and the truncated protein was incorporated into IVs and MVs (12). Since signal peptidase is located within the lumen of the ER, these data provided evidence for an operative pathway between the ER and the viral membrane. Furthermore, we showed that replacing the cytoplasmic tail (CT) of A9 with one containing COPII-binding sites diverted it to the Golgi apparatus, consistent with initial ER insertion (12). The purpose of the present study was to determine the putative signals that direct viral transmembrane proteins to the nascent IV membrane.
MATERIALS AND METHODS
Plasmids.
Most of the plasmids used for transfection were derivatives of pGEM7 (Promega, Madison, WI) constructed by PCR. Plasmids pcTM-A9 and pcTM-A9(CT-VG) were regulated by the cytomegalovirus early promoter for expression in uninfected cells. Those plasmids used for expression in infected cells, namely, pGA9, pGA9ΔNT, pGA9ΔTM, pGA9ΔtTM, pGA9ΔCT, pGNT-A9, pGTM-A9, pGtTM-A9, pGCT-A9, pGA9(TM-B5), pGA9(TM-VG), pGTM-B5, and pGTM-VG, were regulated by the VACV A9L promoter.
Antibodies.
Anti-HA.11 mouse monoclonal antibody (MAb) and rabbit polyclonal antibody, which recognize the influenza hemagglutinin (HA) epitope, were purchased from Covance (Princeton, NJ). Rabbit anti-A17LC polyclonal antibody, which recognizes the C-terminal peptide of the A17 protein, was described elsewhere (37). Anti-B5 rat MAb 192C (24) was prepared from a hybridoma provided by J. Locker (European Molecular Biology Laboratories, Heidelberg, Germany). Rabbit anti-β-COP polyclonal antibody and a mouse protein disulfide isomerase (PDI) MAb were purchased from BD Biosciences (San Jose, CA) and Stressgen Bioreagents (Ann Arbor, MI), respectively. Alexa 488-conjugated anti-mouse immunoglobulin G (IgG) and anti-rabbit IgG, Alexa 594-conjugated anti-mouse IgG, Alexa 568-conjugated anti-rat IgG, and 4′,6′-diamidino-2-phenylindole (DAPI) were purchased from Invitrogen (Carlsbad, CA). Rhodamine red-conjugated anti-rabbit IgG antibody was purchased from Jackson Immunoresearch Laboratories (West Grove, PA).
Transfection and infection.
Lipofectamine 2000 (Invitrogen) and DNA were diluted separately in Opti-MEM I medium (Invitrogen), mixed, incubated at room temperature for 20 min, and added to HeLa cells for 4 to 5 h at 37°C. The Lipofectamine-DNA medium was replaced with fresh Dulbecco's modified Eagle's medium from Quality Biologicals (Gaithersburg, MD), supplemented with 10% fetal bovine serum (FBS). For infection, virus stocks diluted in Dulbecco's modified Eagle's medium supplemented with 2.5% FBS were added to cell monolayers. After 1 h at 37°C, the virus inoculum was replaced with fresh medium containing 2.5% FBS.
Metabolic labeling and immunoprecipitation.
HeLa cells were starved for 30 min at 37°C in methionine- and cysteine-free minimum essential medium without serum and then labeled with 100 to 200 μCi per ml of an [35S]methionine and [35S]cysteine mixture in the above medium for 30 to 120 min. Cells were harvested in cold phosphate-buffered saline (PBS) and lysed immediately in ice-cold radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate). Lysates were incubated on ice for 10 min and centrifuged at 20,000 × g for 10 min at 4°C. Antibody was added to the supernatant and incubated overnight at 4°C. On the next day, protein G-agarose (Roche Applied Sciences, Indianapolis, IN) was added to each lysate and incubated as described above for 2 h. Agarose beads were pelleted at 20,000 × g for 30 s at 4°C and then washed four times with RIPA buffer and once with PBS. Lithium dodecyl sulfate sample buffer (Invitrogen) was added to agarose beads, and proteins were resolved in 12% bis-Tris polyacrylamide gels with 2-morpholinoethanesulfonic acid buffer (Invitrogen) and visualized by autoradiography. Films were scanned and images were compiled with Adobe (San Jose, CA) Photoshop, version 7.0.1, software.
Confocal microscopy.
Cells were washed with PBS and fixed with cold 4% paraformaldehyde in PBS at room temperature for 20 min. Fixed cells were treated for 5 min with 0.2% Triton X-100 in PBS at room temperature or with 20 μg/ml of digitonin in PBS at 0°C. Permeabilized cells were incubated with primary antibodies diluted in 10% FBS for 1 h, followed by secondary antibody diluted in 10% FBS for 30 min, at room temperature. For double staining, cells were incubated sequentially with each primary and secondary antibody and washed at least three times with PBS after incubation with each antibody. Finally, cells were stained with DAPI diluted in PBS (5 to 10 μg/ml) for 10 min at room temperature. Stained cells were washed extensively with PBS, and coverslips were mounted in 20% glycerol. Fluorescence was examined with a 63×/1.4 oil immersion objective attached to a Leica inverted confocal microscope, and images were collected using Leica confocal SP2 software (Leica Microsystems, Heidelberg, Germany). Photos were processed using Adobe Photoshop, version 7.0.1, software.
Transmission electron microscopy of immunogold-labeled thawed cryosections.
Infected cells were washed, fixed with 4% paraformaldehyde-0.05% glutaraldehyde, impregnated with 2.3 M sucrose, quick-frozen, and cut into 70-nm-thick sections. Cryosections were picked up on grids, thawed, washed free of sucrose, and stained with a mouse MAb to the HA epitope tag, followed by rabbit anti-mouse IgG from Cappel-ICN Pharmaceuticals (Aurora, OH) and then protein A conjugated to 10-nm gold spheres (University Medical Center Utrecht, Utrecht, The Netherlands). The sections were analyzed on a CM100 transmission electron microscope (FEI, Hillsboro, OR).
RESULTS
Construction and expression of a panel of mutated A9 proteins.
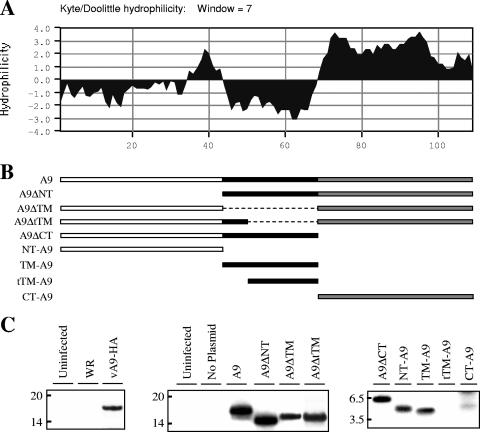
VACV A9, a nonglycosylated protein with a predicted mass of 12 kDa, can be divided into a moderately hydrophobic N-terminal region (NT), a central transmembrane domain (TM), and a hydrophilic C-terminal CT (Fig. 1A). To identify putative signals that target A9 to viral membranes, we cloned the A9 open reading frame with a deletion of amino acids 2 to 43, comprising the NT (A9ΔNT), 44 to 68, comprising the entire putative TM (A9ΔTM), 52 to 68, comprising part of the TM (A9ΔtTM), and 69 to 108, comprising the CT (A9ΔCT) (Fig. 1B). In addition, the NT (NT-A9), TM (TM-A9), partial TM (tTM-A9), and CT (CT-A9) were cloned into plasmids (Fig. 1B). Each modified gene retained the natural A9 promoter so that mRNA synthesis would occur in the virus factory by the VACV transcription system. In addition, the encoded proteins had a C-terminal HA tag allowing detection by a MAb. Cells were infected with a conditionally lethal mutant, vA9i (38), in the absence of inducer in order to prevent expression of the untagged A9 protein from the viral genome, which could potentially compete with A9 expressed from a transfected plasmid. Following infection and transfection, the cells were labeled with [35S]methionine, and HA-tagged proteins were captured with a MAb and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The left panel of Fig. 1C shows negative controls from uninfected cells and cells infected with wild-type VACV strain WR without an HA epitope tag and a positive control from cells infected with a recombinant VACV that expresses A9-HA. The middle and right panels of Fig. 1C show labeled proteins from cells infected with vA9i in the absence of inducer and transfected with plasmids containing the constructs shown in Fig. 1B. The proteins all appeared to be stably synthesized, except for tTMA9, which was not detected here (Fig. 1C) and was demonstrated to be unstable in pulse-chase experiments (not shown). The level of CT-A9 appeared to be reduced, presumably because of the presence of only one methionine that could be 35S labeled.
FIG. 1.
Construction and expression of A9 with single or multiple deletions. (A) Kyte-Doolittle hydrophilicity plot of A9. (B) Diagrams of A9 segments used to construct plasmids are shown, with plasmid names at the left. Dashed lines represent deleted DNA. (C) Synthesis of full-length and truncated A9 proteins. (Left panel) HeLa cells were uninfected or infected with VACV (strain WR) or vA9-HA expressing HA-tagged A9. (Middle and right panels) Cells were uninfected or infected with vA9i and transfected with the indicated plasmids. Sixteen hours after infection, the cells were labeled with [35S]methionine-cysteine. Cleared lysates were immunoprecipitated with anti-HA MAb, and the bound proteins were resolved by SDS-polyacrylamide gel electrophoresis and visualized by autoradiography. Some samples were run in separate gels and combined for presentation.
Intracellular location of A9 is independent of the N- or C-terminal domain.
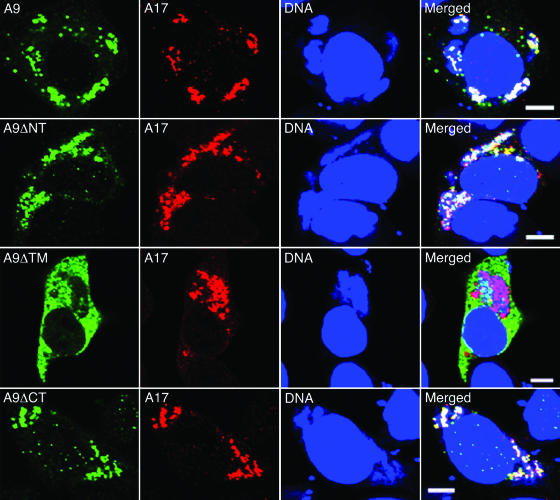
Assembly of the viral membrane occurs in cytoplasmic factories that can be recognized by DNA staining with DAPI and distinguished from the adjacent nucleus by their smaller size, less regular shape, and antibody reactivity to viral proteins. Full-length A9 largely colocalized with A17, a protein component of the IV membrane that is essential for its formation (21, 37), within DAPI-staining DNA factories (Fig. 2, row 1). Both the N-terminally deleted (Fig. 2, row 2) and C-terminally deleted (Fig. 2, row 4) forms of A9 also colocalized with A17 in factories, indicating that these sequences were not required for proper intracellular distribution. In contrast, the A9 protein missing the TM (Fig. 2, row 3) or part of the TM (not shown) exhibited diffuse cytoplasmic staining and was largely excluded from the factory region.
FIG. 2.
Effects of deletions on intracellular location of A9. HeLa cells infected with vA9i and transfected with a plasmid expressing full-length A9 or A9 with an N-terminal (A9ΔNT), TM (A9ΔTM), or C-terminal (A9ΔCT) deletion were fixed and permeabilized 16 h after infection. Cells were stained with mouse anti-HA MAb followed by Alexa 488-conjugated anti-mouse IgG and then with rabbit anti-A17 polyclonal antibody followed by rhodamine red-conjugated anti-rabbit IgG. Finally, cells were stained with DAPI and visualized by confocal microscopy. Green, Alexa 488; red, rhodamine red; blue, DAPI. Bars, 10 μm.
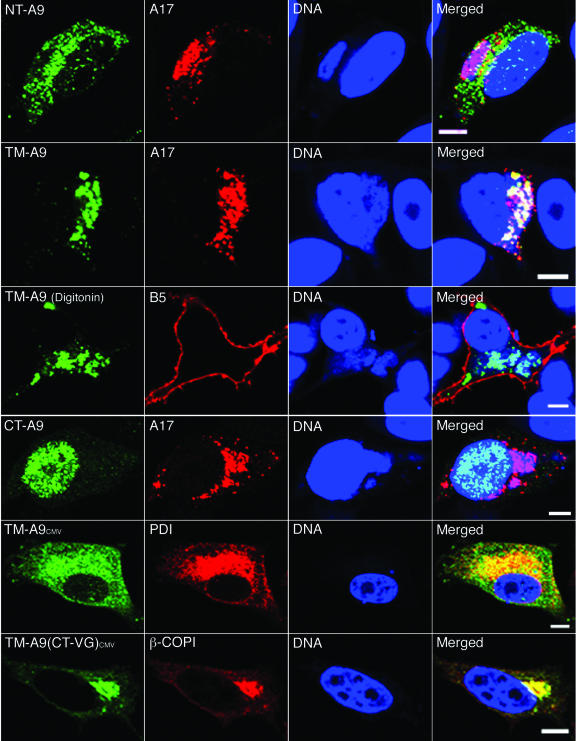
The above analysis indicated that only the TM was essential for localization of the A9 protein in viral factories. Further analysis demonstrated that the A9 TM without additional sequences except for the HA epitope tag was sufficient for colocalization with A17 in the factory (Fig. 3, row 2), whereas the NT (Fig. 3, row 1) and CT (Fig. 3, row 4) domains were insufficient. The nuclear localization of the CT protein was presumably due to short positively charged amino acid sequences. Analysis of cells treated with digitonin, which permeabilizes the plasma membrane and allows penetration of the anti-HA MAb, indicated that the C terminus of the A9 TM protein is in the cytoplasm (Fig. 3, row 3), just like the case with full-length A9 (12). The specificity of digitonin permeabilization was confirmed by detection of the luminal domain of the B5 protein only on the cell surface, not in the ER or Golgi apparatus (Fig. 3, row 3).
FIG. 3.
Intracellular locations of polypeptides containing the NT, TM, or CT of A9. HeLa cells were infected with vA9i and transfected with a plasmid expressing the HA epitope-tagged A9 NT (NT-A9; row 1), TM (TM-A9; rows 2 and 3), or CT (CT-A9; row 4) from the natural A9 promoter or with a plasmid expressing TM-A9 (row 5) or TM-A9 with the VSV G CT [TM-A9(CT-VG); row 6] from a cytomegalovirus promoter. After 16 h (rows 1 to 4) or 24 h (rows 5 and 6), cells were fixed and permeabilized with Triton X-100 (rows 1, 2, 4, 5, and 6) or digitonin (row 3). Cells were stained with mouse anti-HA MAb followed by Alexa 488-conjugated anti-mouse IgG (rows 1, 2, 3, 4, and 6) or with rabbit anti-HA polyclonal antibody followed by Alexa 488-conjugated anti-rabbit IgG (row 5). Cells were then stained with rabbit anti-A17 polyclonal antibody followed by rhodamine red-conjugated anti-rabbit IgG (rows 1, 2, and 4), rat anti-B5 MAb followed by Alexa 568-conjugated anti-rat IgG (row 3), mouse anti-PDI MAb followed by Alexa 594-conjugated anti-mouse IgG (row 5), or rabbit anti-β-COPI polyclonal antibody followed by rhodamine red-conjugated anti-rabbit IgG (row 6). Finally, nuclei and viral factories were stained with DAPI and visualized by confocal microscopy. Green, Alexa 488; red, rhodamine red, Alexa 568, and Alexa 594; blue, DAPI. Bars, 10 μm.
It was also of interest to determine the localization of the A9 TM protein in uninfected cells. This was accomplished by using the cytomegalovirus RNA polymerase II promoter for transcription of the transfected A9 TM gene. The location of this protein overlapped with that of PDI, an intraluminal ER protein (Fig. 3, row 5). In addition, when the CT of the vesicular stomatitis virus (VSV) G protein was appended to the A9 TM protein, the chimeric protein localized mostly with the β-COP Golgi membrane marker (Fig. 3, row 6), similar to the result previously determined for both infected and uninfected cells by replacing the CT of the full-length A9 protein with the corresponding region of VSV G (12).
Incorporation of truncated A9 polypeptides in IV and MV membranes.
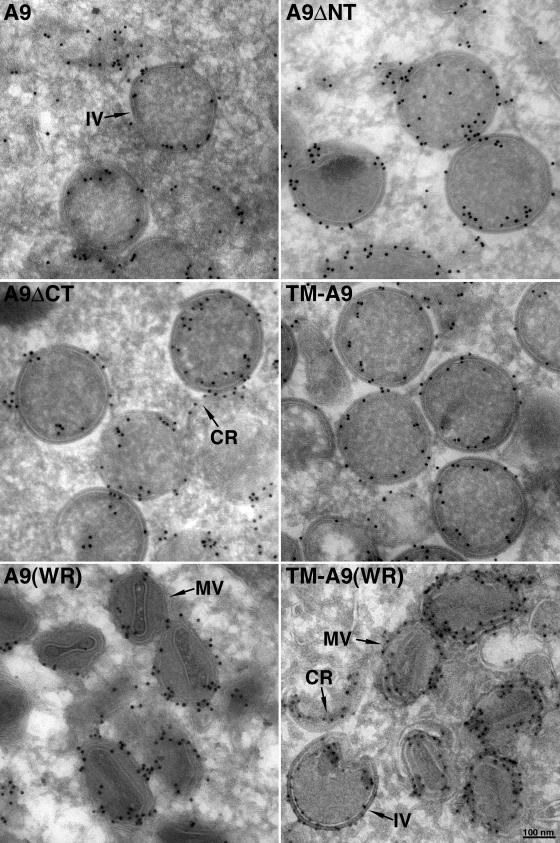
Further experiments demonstrated that truncated forms of A9 that retained the TM were incorporated into IV and MV membranes within cytoplasmic factories. IVs are spherical particles that appear on cross section as circular structures bounded by a membrane and an external lattice, whose apparent thickness depends on the level of the cross section relative to the equator (33). We followed the basic protocol used in the preceding sections for confocal microscopy by infecting cells with vA9i in the absence of inducer and transfecting them with a plasmid expressing A9, A9ΔNT, A9ΔTM, A9ΔCT, or TM-A9. The cells were then cryosectioned, and the HA epitope was detected by immunogold staining. In each case, except for A9ΔTM, gold particles were found at the membrane near the circumference of the IV (Fig. 4, rows 1 and 2). In the cells expressing A9ΔTM, the gold grains were scattered and not associated with distinct structures (not shown), as anticipated from confocal microscopy (Fig. 2, row 3). Despite the association of the A9 protein lacking both the NT and CT with IV membranes, the protein did not complement the production of infectious virus particles, whereas A9 missing only the CT was able to do so (12; M. Husain, unpublished data). These data suggest that the N-terminal domain is required for virus morphogenesis rather than membrane insertion and that the CT is dispensable.
FIG. 4.
Immunogold electron microscopy showing A9 or mutated A9 proteins incorporated into IV and MV membranes. HeLa cells were infected with vA9i and transfected with a plasmid encoding A9 or A9 missing the NT (A9ΔNT), the CT (A9ΔCT), or both the NT and CT (TM-A9) (rows 1 and 2) or infected with wild-type VACV strain Western Reserve (WR) and transfected with A9 or TM-A9 (row 3). After 16 h (rows 1 and 2) or 24 h (row 3), cells were fixed, cryosectioned, and stained with anti-HA MAb followed by rabbit anti-mouse IgG and protein A conjugated to 10-nm gold spheres. IVs and a crescent (CR) are shown in rows 1 and 2, and MVs are shown in row 3. In the right panel of row 3, there is a crescent and an IV in the lower left section.
To take the analysis a step further, we infected cells with wild-type VACV strain WR so that full-length A9 would be made, allowing the formation of MVs as well as IVs. Following transfection, HA-tagged TM-A9 as well as full-length A9 was detected in MVs, which were recognized by their oval shape and internal core (Fig. 4, row 3). This result confirms the insertion of the epitope-tagged TM protein into bona fide viral membranes.
Protein targeting to viral membranes does not depend on specific amino acid sequences.
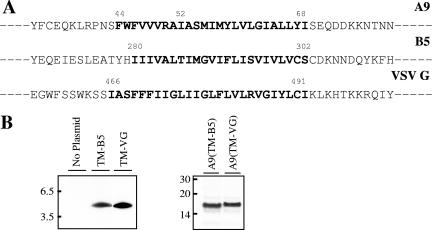
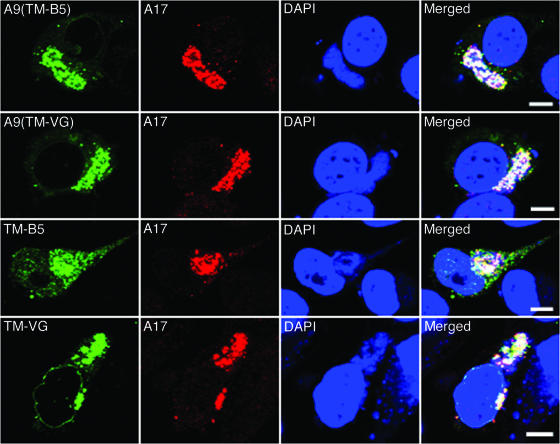
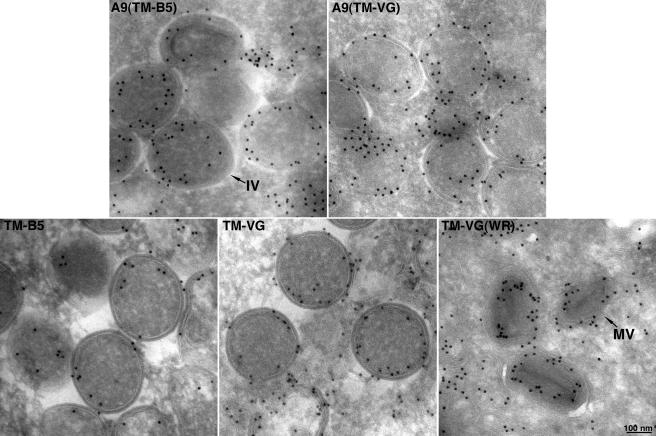
Since neither the NT nor CT of A9 is required for incorporation of the latter into the viral membrane, we considered that the TM might have a specific targeting sequence. Upon inspection of the corresponding regions of A9 orthologs in other poxviruses, however, we found that only Gly at position 62 of A9 was conserved. However, when this Gly residue was changed to Arg, the viral factory distribution of the mutated A9 protein was no different from that of wild-type A9 (data not shown), suggesting no absolute sequence requirement for targeting. The above result raised the possibility that any TM could enable viral membrane localization. To evaluate this hypothesis, the putative TM of A9 was replaced with the unrelated TM of the VACV B5 protein (Fig. 5A), which traffics to the trans-Golgi and EV membranes in infected cells, or with the TM domain of the VSV G protein (Fig. 5A), which traffics to Golgi and plasma membranes. In addition, we expressed these heterologous TMs with a C-terminal HA tag without any A9 coding sequence. Synthesis of each protein was detected by immunoprecipitation and SDS-polyacrylamide gel electrophoresis for cells infected with vA9i and transfected with the appropriate plasmid (Fig. 5B). We found by confocal microscopy that the A9 proteins containing the VACV B5 or VSV G TM instead of the A9 TM still colocalized with the VACV A17 protein in viral factories (Fig. 6, rows 1 and 2). Furthermore, the TMs of VACV B5 and VSV G lacking A9 sequences also colocalized with the A17 protein (Fig. 6, rows 3 and 4). Although a chimera consisting of the B5 TM appended to the VSV G luminal domain was previously reported to localize in Golgi membranes of uninfected cells (36), we did not observe any tendency of the B5 TM, alone or with A9 sequences, to proceed to the Golgi apparatus in the present constructs. The association of these TM proteins with IV and MV membranes was demonstrated by immunoelectron microscopy (Fig. 7). These data suggest that the apolar nature of the TM, rather than a specific amino acid sequence, suffices for targeting of a protein to viral membranes in factories.
FIG. 5.
Heterologous TM constructs. (A) Partial amino acid sequences of VACV A9 and B5 and of VSV G, with their TMs shown in bold. (B) Synthesis of B5 and VSV G TM polypeptides or of A9 with the B5 or VSV G TM. HeLa cells were infected with vA9i and transfected with the indicated plasmids. Sixteen hours after infection, cells were labeled with [35S]methionine-cysteine. Cleared lysates were immunoprecipitated with anti-HA MAb, and bound proteins were analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography.
FIG. 6.
Localization of polypeptides with heterologous TMs. HeLa cells were infected with vA9i and transfected with plasmid A9(TM-B5) or A9(TM-VG), encoding A9 with the TM of B5 or VSV G, respectively, or with TM-B5 or TM-VG, encoding the TM alone of B5 or VSV G, respectively. In each case, the gene had an A9 promoter and the protein had a C-terminal HA epitope tag. Cells were stained with mouse anti-HA MAb followed by Alexa 488-conjugated anti-mouse IgG. Cells were then stained with rabbit anti-A17 polyclonal antibody followed by rhodamine red-conjugated anti-rabbit IgG. Finally, nuclei and viral factories were stained with DAPI and visualized with a confocal microscope. Green, Alexa 488; red, rhodamine red; blue, DAPI. Bars, 10 μm.
FIG. 7.
Immunogold electron microscopy of polypeptides with heterologous TMs. Infections with vA9i and transfections were carried out as described in the legend to Fig. 6, except for the rightmost panel of row 2, in which cells were infected with VACV WR. Immunoelectron microscopy was done as described in the legend to Fig. 4. The rightmost panel in row 2 contains MVs; all other panels contain IVs.
DISCUSSION
To learn more about the formation of the poxvirus membrane, we analyzed the trafficking of VACV A9, a conserved MV protein that is essential for morphogenesis. We previously demonstrated that VACV A9 inserts cotranslationally into microsomes in vitro, localizes within the ER when expressed in uninfected cells, and is found in IV membranes and nearby tubules containing the ER luminal protein PDI in infected cells (12). Cleavage of a heterologous signal peptide, which replaced the NT of A9, and incorporation of the truncated protein into IV and MV membranes provided compelling evidence for an operative pathway between the secretory system of the cell and the viral membrane (12). The inability of a dominant-negative SarI GTPase, which blocks the COPII ER exit pathway, to block IV and MV formation suggested that trafficking could occur directly from the ER (11).
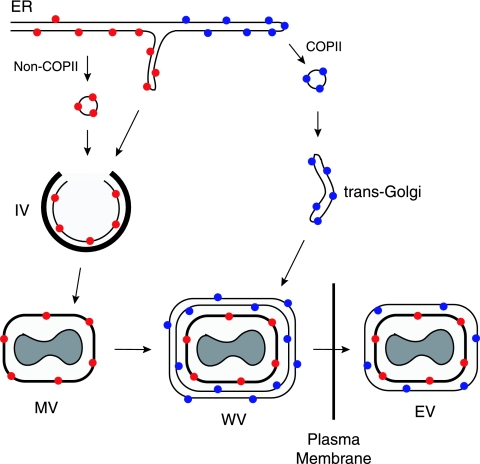
The purpose of the present study was to determine protein sequences required for the localization of proteins in the nascent viral membrane. To our surprise, neither the N- nor C-terminal domain of A9 was needed. Furthermore, a heterologous TM from the VSV G protein or the VACV B5 EV protein could replace the A9 TM, indicating either a complete lack of sequence specificity or redundant signals in the N- and C-terminal domains such that deletion of either one had no effect. The latter possibility was ruled out by showing that the A9 TM or a heterologous EV or nonpoxvirus TM was sufficient for viral membrane targeting. These data imply a default pathway for the localization of TM proteins in the viral membrane. However, such a model seemed at odds with the knowledge that only specific viral proteins are directed to the IV membrane, while others continue on the secretory pathway to the Golgi apparatus. How can these seemingly disparate data be reconciled? One clue came from our observation that replacing the CT of A9 with the corresponding region of the VSV G protein, which contains COPII ER exit site binding motifs (17, 18, 26), diverts ER-viral membrane trafficking to the Golgi apparatus (12). The addition of a long luminal domain, which could lead to interactions with ER-resident proteins, also interfered with IV membrane localization (Husain, unpublished data), perhaps explaining the absence of glycosylated viral proteins in virions. Indeed, most of the ∼20 MV membrane proteins have a very short or even no luminal domain (2). The A14 protein is exceptional in that it has two membrane-spanning domains with N and C termini in the lumen (15). Although glycosylated A14 can be detected in infected cells, it is not packaged in virions, and the amount of the glycosylated protein is increased under conditions that interfere with virus membrane formation (15, 19). Glycosylation and small amounts of an MV protein in the ERGIC may represent escape from the biologically relevant ER-IV trafficking pathway. A model depicting the synthesis of proteins without and with COPII-binding motifs and destined for the IV and EV membranes, respectively, is shown in Fig. 8.
FIG. 8.
Sorting of proteins to MV and EV membranes. Membrane proteins synthesized in viral factory areas are translocated into the ER and sorted based on the presence (blue dots) or absence (red dots) of COPII ER exit signals in their CTs. ER patches containing unglycosylated proteins lacking ER exit signals or other sequences that interact with ER-resident proteins form vesicles or tubules that fuse with viral crescent membranes containing an external protein scaffold (thick black line) to form IVs. IVs lose the scaffold and undergo further morphogenesis to form MVs. Proteins with ER exit signals traverse the exocytic pathway to the trans-Golgi, where cisternal elements wrap the MVs in a double membrane. The wrapped virions (WVs) move along microtubules and fuse with the plasma membrane to release EVs with one additional membrane relative to MVs.
If trafficking of proteins without conventional ER exit signals to the IV membrane is a default pathway, then why is there no evidence of cellular proteins in IV and MV membranes? We suspect that the absence of cellular proteins results from the specialized site of viral protein synthesis. We will show elsewhere (G. Katsafanas and B. Moss, unpublished data) that the presence of ribosomes and translation initiation factors within the virus factory allows the coordination of viral transcription and translation. Since it appears that only ER domains within the virus factory are remodeled into viral membranes, only proteins synthesized there may have the opportunity to associate with the nascent viral membrane. The nearly complete shutdown of host protein synthesis at late times after infection may also contribute to the exclusion of nonviral proteins. The latter feature, moreover, makes it difficult to determine whether proteins made outside the factory could be incorporated into viral membranes. We tried to get around this problem by expressing an epitope-tagged A9 protein just prior to infection and found that the protein remained in the ER outside the factory area, as predicted (Husain, unpublished data).
There are intriguing similarities between the formation of peroxisome and poxvirus membranes. Peroxisomes are discrete organelles in the cytoplasm of eukaryotic cells. Their ability to regenerate in cells apparently lacking them led to the now disfavored hypothesis that peroxisomal membranes form de novo (32). The finding that peroxisome formation occurs in the presence of a constitutively inactive Sar1 inhibitor indicated that COPII trafficking was not involved (31), similar to the result obtained for VACV IV formation (11). Moreover, vesicles or tubules from the ER contribute to peroxisome formation even though ER proteins are excluded from these organelles (9, 34), as is the case with VACV. Microscopic studies indicated that a peroxisomal TM protein called Pex3 concentrates in foci in the ER and is joined by a farnesylated cytosolic protein called Pex19, both of which are necessary for peroxisome formation. Subsequently, many other proteins are recruited to the peroxisome via a conserved sequence (23), which has no parallel in VACV proteins. Nevertheless, whether related mechanisms underlie peroxisome and poxvirus membrane formation remains to be determined.
Acknowledgments
We thank Victor Hsu, Ted Pierson, and Jon Yewdell for comments.
The intramural program of NIAID, NIH, supported this study.
Footnotes
Published ahead of print on 27 December 2006.
REFERENCES
- 1.Carter, G. C., M. Law, M. Hollinshead, and G. L. Smith. 2005. Entry of the vaccinia virus intracellular mature virion and its interactions with glycosaminoglycans. J. Gen. Virol. 86:1279-1290. [DOI] [PubMed] [Google Scholar]
- 2.Condit, R. C., N. Moussatche, and P. Traktman. 2006. In a nutshell: structure and assembly of the vaccinia virion. Adv. Virus Res. 66:31-124. [DOI] [PubMed] [Google Scholar]
- 3.Cyrklaff, M., C. Risco, J. J. Fernandez, M. V. Jimenez, M. Esteban, W. Baumeister, and J. L. Carrascosa. 2005. Cryo-electron tomography of vaccinia virus. Proc. Natl. Acad. Sci. USA 102:2772-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dales, S., and E. H. Mosbach. 1968. Vaccinia as a model for membrane biogenesis. Virology 35:564-583. [DOI] [PubMed] [Google Scholar]
- 5.Ericsson, M., B. Sodeik, J. K. Locker, and G. Griffiths. 1997. In vitro reconstitution of an intermediate assembly stage of vaccinia virus. Virology 235:218-227. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths, G., N. Roos, S. Schleich, and J. K. Locker. 2001. Structure and assembly of intracellular mature vaccinia virus: thin-section analyses. J. Virol. 75:11056-11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grimley, P. M., E. N. Rosenblum, S. J. Mims, and B. Moss. 1970. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J. Virol. 6:519-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heuser, J. 2005. Deep-etch EM reveals that the early poxvirus envelope is a single membrane bilayer stabilized by a geodetic “honeycomb” surface coat. J. Cell Biol. 169:269-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoepfner, D., D. Schildknegt, I. Braakman, P. Philippsen, and H. F. Tabak. 2005. Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122:85-95. [DOI] [PubMed] [Google Scholar]
- 10.Hollinshead, M., A. Vanderplasschen, G. L. Smith, and D. J. Vaux. 1999. Vaccinia virus intracellular mature virions contain only one lipid membrane. J. Virol. 73:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Husain, M., and B. Moss. 2003. Evidence against an essential role of COPII-mediated cargo transport to the endoplasmic reticulum-Golgi intermediate compartment in the formation of the primary membrane of vaccinia virus. J. Virol. 77:11754-11766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Husain, M., A. S. Weisberg, and B. Moss. 2006. Existence of an operative pathway from the endoplasmic reticulum to the immature poxvirus membrane. Proc. Natl. Acad. Sci. USA 103:19506-19511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krijnse-Locker, J., S. Schleich, D. Rodriguez, B. Goud, E. J. Snijder, and G. Griffiths. 1996. The role of a 21-kDa viral membrane protein in the assembly of vaccinia virus from the intermediate compartment. J. Biol. Chem. 271:14950-14958. [DOI] [PubMed] [Google Scholar]
- 14.Law, M., G. C. Carter, K. L. Roberts, M. Hollinshead, and G. L. Smith. 2006. Ligand-induced and non-fusogenic dissolution of a viral membrane. Proc. Natl. Acad. Sci. USA 103:5989-5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer, J., and P. Traktman. 2003. Investigation of structural and functional motifs within the vaccinia virus A14 phosphoprotein, an essential component of the virion membrane. J. Virol. 77:8857-8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moss, B. 2006. Poxvirus entry and membrane fusion. Virology 344:48-54. [DOI] [PubMed] [Google Scholar]
- 17.Nishimura, N., and W. E. Balch. 1997. A di-acidic signal required for selective export from the endoplasmic reticulum. Science 277:556-558. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura, N., S. Bannykh, S. Slabough, J. Matteson, Y. Altschuler, K. Hahn, and W. E. Balch. 1999. A di-acidic (DXE) code directs concentration of cargo during export from the endoplasmic reticulum. J. Biol. Chem. 274:15937-15946. [DOI] [PubMed] [Google Scholar]
- 19.Resch, W., A. S. Weisberg, and B. Moss. 2005. Vaccinia virus nonstructural protein encoded by the A11R gene is required for formation of the virion membrane. J. Virol. 79:6598-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Risco, C., J. R. Rodriguez, C. Lopez-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodríguez, D., M. Esteban, and J. R. Rodríguez. 1995. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J. Virol. 69:4640-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmons, T., A. Kuhn, F. Wylie, S. Schleich, J. R. Rodriguez, D. Rodriguez, M. Estban, G. Griffiths, and J. K. Locker. 1997. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J. Virol. 71:7404-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schliebs, W., and W. H. Kunau. 2004. Peroxisome membrane biogenesis: the stage is set. Curr. Biol. 14:R397-R399. [DOI] [PubMed] [Google Scholar]
- 24.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans-Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Senkevich, T. G., S. Ojeda, A. Townsley, G. E. Nelson, and B. Moss. 2005. Poxvirus multiprotein entry-fusion complex. Proc. Natl. Acad. Sci. USA 102:18572-18577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sevier, C. S., O. A. Weisz, M. Davis, and C. E. Machamer. 2000. Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol. Biol. Cell 11:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, G. L., A. Vanderplasschen, and M. Law. 2002. The formation and function of extracellular enveloped vaccinia virus. J. Gen. Virol. 83:2915-2931. [DOI] [PubMed] [Google Scholar]
- 28.Sodeik, B., S. Cudmore, M. Ericsson, M. Esteban, E. G. Niles, and G. Griffiths. 1995. Assembly of vaccinia virus: incorporation of p14 and p32 into the membrane of the intracellular mature virus. J. Virol. 69:3560-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sodeik, B., G. Griffiths, M. Ericsson, B. Moss, and R. W. Doms. 1994. Assembly of vaccinia virus: effects of rifampin on the intracellular distribution of viral protein p65. J. Virol. 68:1103-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.South, S. T., K. A. Sacksteder, X. Li, Y. Liu, and S. J. Gould. 2000. Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J. Cell Biol. 149:1345-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramani, S. 1998. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol. Rev. 78:171-188. [DOI] [PubMed] [Google Scholar]
- 33.Szajner, P., A. S. Weisberg, J. Lebowitz, J. Heuser, and B. Moss. 2005. External scaffold of spherical immature poxvirus particles is made of protein trimers, forming a honeycomb lattice. J. Cell Biol. 170:971-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tabak, H. F., J. L. Murk, I. Braakman, and H. J. Geuze. 2003. Peroxisomes start their life in the endoplasmic reticulum. Traffic 4:512-518. [DOI] [PubMed] [Google Scholar]
- 35.Townsley, A. C., A. S. Weisberg, T. R. Wagenaar, and B. Moss. 2006. Vaccinia virus entry into cells via a low pH-dependent-endosomal pathway. J. Virol. 80:8899-8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward, B. M., and B. Moss. 2000. Golgi network targeting and plasma membrane internalization signals in vaccinia virus B5R envelope protein. J. Virol. 74:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolffe, E. J., D. M. Moore, P. J. Peters, and B. Moss. 1996. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J. Virol. 70:2797-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yeh, W. W., B. Moss, and E. J. Wolffe. 2000. The vaccinia virus A9L gene encodes a membrane protein required for an early step in virion morphogenesis. J. Virol. 74:9701-9711. [DOI] [PMC free article] [PubMed] [Google Scholar]