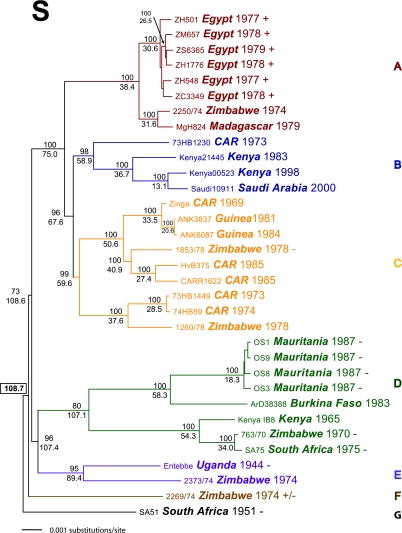
FIG. 2.
Thirty-three complete RVF S segments were analyzed by either unconstrained ML techniques with 500 replicate bootstrap values (PAUP*b10) or the Bayesian statistical program (BEAST) with an MCMC chain length of 3.0 × 107, a 3.0 × 106 burn in, a GTR+Γ+I nucleotide rate substitution model, a strict molecular clock, and sampling of every 1,000 states. A strict molecular clock was chosen after multiple analyses; using both relaxed exponential and relaxed logarithmic clock models revealed no significant deviation in the coalescence of the overall evolutionary rate or tree topologies. Trees and support values from the ML and BEAST analyses were similar. The Bayesian MAP log likelihood value tree was chosen from the posterior distribution and is depicted here. Posterior support values were calculated from consensus analysis of all Bayesian posterior trees, with values over 0.5 indicated above each node. The estimated TMRCA of the tree nodes was calculated by Bayesian analyses and is reported below each node as years prior to the year 2000. The overall TMRCA of the entire S segment analysis is indicated in a text box located adjacent to the root node of SA51. Each taxon name indicates the strain, country of origin, and date of isolation. The GenBank accession numbers for the virus S segments are DQ380143 to -6, DQ380149, DQ380151 to -3, DQ380156, and DQ380158 to -81. Strains used in previous studies of virulence in WF rats are indicated with either + (lethal; LD50, ∼1.0 PFU), +/−, (lethal; LD50, ∼2 × 103 PFU), or − (nonlethal).