FIG. 5.
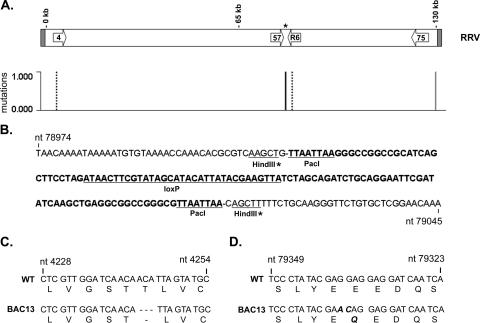
CGH analysis and identification of mutations in the RRV BAC and BAC-derived viral isolate DNA. RRV BAC DNA and BAC-derived RRV isolate 13 viral DNA were used in CGH analysis with RRV17577 as the reference genome. Differences in genomic sequence were identified based on variations in hybridization intensities, indicating locations of potential mutations in the RRV sequence (A). The mutation found only in the RRV BAC is indicated by a solid gray line, mutations found only in BAC-derived isolate 13 are indicated by a dashed black line, and the common mutation found in both samples is indicated by a solid black line. PCR of the region encompassing the BAC vector insertion was performed using BAC-derived RRV isolate 13, and the resulting product was directly sequenced. (B) The sequence indicates that the mutation identified near nt 79010 is due to the presence of 120 bp of pKSO-gpt sequence at the site of the BAC vector insertion that remains after BAC vector removal by CRE recombinase. The pKSO-gpt sequence is indicated in bold type, with the PacI sites used for cloning and the loxP site underlined. The partial HindIII sites remaining after the insertion of pKSO-gpt into RRV are also noted, with an asterisk. PCR was also performed using BAC-derived RRV isolate 13 as a template to amplify regions encoding ORF4 and R6, and the resulting products were directly sequenced. The sequence alterations detected include a 3-bp deletion in the ORF4 coding sequence (nt 4243 to 4245), which results in the deletion of Thr190 from the vCCP encoded by ORF4 (C) and the conversion of nt 79338 and 79337 in the R6 coding sequence from GG to AC, ultimately causing the conversion of Glu393 to Gln in the encoded vIRF (D).