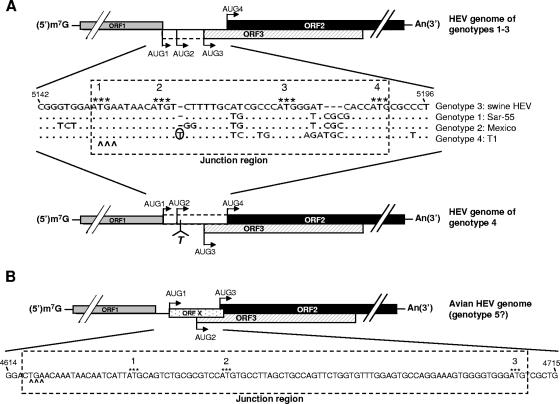
FIG. 1.
Schematic diagram of the predicted genomic organization and nucleotide sequence alignment of the noncoding junction region between ORF1 and ORF2 of HEV. (A) Swine HEV (genotype 3), Sar-55 strain (genotype 1), Mexico strain (genotype 2), and T1 strain (genotype 4). Nucleotide position numbers correspond to the sequence of clone pSHEV-3 of genotype 3 swine HEV. Dots denote sequence identity, and dashes denote deletions. The stop codon of ORF1 is indicated by carets, and the four AUGs are indicated by asterisks. AUG1, -2, and -3 are in-frame and serve as the potential ORF3 start codons, and AUG4 serves as the ORF2 start codon among HEV genotypes 1 to 3. In genotype 4, a single-nucleotide T insertion 2 nt downstream of AUG2 (marked with circle) results in the predicted ORF3 initiation at AUG3 and the predicted ORF2 initiation at AUG1, -2, and/or -4. The presumed coding regions between AUG1 and AUG3 for ORF3 in HEV genotypes 1 to 3 and between AUG1 and AUG4 for ORF2 in genotype 4 HEV are denoted by the dashed-line box based on our hypothesis that AUG3 is the authentic ORF3 initiation site and that AUG4 is the authentic ORF2 initiation site for all HEV genotypes. (B) Avian HEV (putative genotype 5). There exist three AUGs that could potentially serve as the initiation sites for different ORFs. In addition to AUG2 and AUG3 as the potential ORF3 and ORF2 start codons, respectively, a novel small ORF X is predicted to start at AUG1.