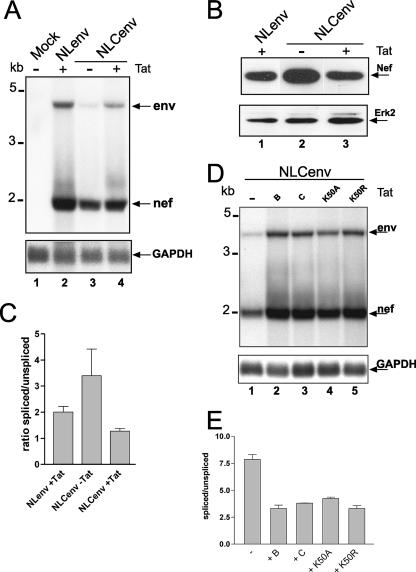
FIG. 2.
Cotransfection of Tat led to reversal of enhanced splicing. (A) LTR- or CMV-containing NLenv plasmids were transfected into HeLa P4 cells in the presence or absence of an HIV-Tat-encoding plasmid. Northern blot analysis was performed as described for Fig. 1C. GAPDH serves as a loading control. Molecular size markers are shown on the left, and the RNAs are named on the right. (B) Western blot analysis of cell lysates from the transfection performed as described for panel A. An anti-Nef antibody was used, and the blot was reprobed with an anti-extracellular signal-regulated kinase 2 (Erk2) antibody for a loading control. (C) Quantification of the Northern blot data using a PhosphorImager (Amersham). Mean values represent the results of four independent experiments. (D) The NLCenv construct was transfected together with different Tat plasmids as indicated or left untransfected (−). + B and + C specify the HIV Tat subtypes and K50A and K50R point mutations based on subtype C Tat. GAPDH served as a loading control. Molecular markers and RNA species are indicated as described for panel A. (E) Quantification of the Northern blot data as described for panel C.