Abstract
Emergence of human immunodeficiency virus type 1 (HIV-1) populations that switch or broaden coreceptor usage from CCR5 to CXCR4 is intimately coupled to CD4+ cell depletion and disease progression toward AIDS. To better understand the molecular mechanisms involved in the coreceptor switch, we determined the nucleotide sequences of 253 V1 to V3 env clones from 27 sequential HIV-1 subtype B isolates from four patients with virus populations that switch coreceptor usage. Coreceptor usage of clones from dualtropic R5X4 isolates was characterized experimentally. Sequence analysis revealed that 9% of the clones from CXCR4-using isolates had originated by recombination events between R5 and X4 viruses. The majority (73%) of the recombinants used CXCR4. Furthermore, coreceptor usage of the recombinants was determined by a small region of the envelope, including V3. This is the first report demonstrating that intrapatient recombination between viruses with distinct coreceptor usage occurs frequently. It has been proposed that X4 viruses are more easily suppressed by the immune system than R5 viruses. We hypothesize that recombination between circulating R5 viruses and X4 viruses can result in chimeric viruses with the potential to both evade the immune system and infect CXCR4-expressing cells. The broadening in cell tropism of the viral population to include CXCR4-expressing cells would gradually impair the immune system and eventually allow the X4 population to expand. In conclusion, intrapatient recombination between viruses with distinct coreceptor usage may contribute to the emergence of X4 viruses in later stages of infection.
Human immunodeficiency virus type 1 (HIV-1) is the fastest evolving human pathogen. The accelerated evolution of HIV-1 is a consequence of genetic drift due to the error-prone viral reverse transcriptase (28), immune system-mediated selection leading to high viral turnover (5), and recombination between two virion-associated RNA genomes during reverse transcription (14). Within the infected host, the combination of recombination, selection, and genetic drift gives rise to complex quasispecies populations. It has been estimated that HIV-1 may be subjected to as many as three to nine recombination events per round of replication (25, 51). Recombination may lead to major genome rearrangements and is important in the generation and diversification of subpopulations. Indeed, recombination has been found repeatedly in studies of HIV evolution and genetics. Recombination events between viruses of different subtypes of the major (M) group have resulted in a number of stable circulating recombinant forms (20, 21, 29, 47). Furthermore, recombination between viruses isolated from different anatomical sites from one individual has been reported (19, 34). Establishment of recombinant viruses within an infected individual may lead to serious consequences, for example, due to the rapid spread of drug resistance in the virus population (18) and accelerated progression toward AIDS (26).
HIV-1 enters target cells through interactions between the viral glycoproteins (gp120 and gp41), the cellular receptor CD4, and a coreceptor, most often CCR5 or CXCR4 (1). CCR5-using (R5) viruses are often present in the early phase of infection, whereas CXCR4-using (X4) viruses usually appear (or become detectable) only at later stages. The broadening of coreceptor usage to include CXCR4 is associated with accelerated loss of CD4 cells and faster progression to AIDS (41). After the appearance of X4 viruses, the R5 and X4 populations most often coexist in the host. The cellular and molecular mechanisms responsible for virus coreceptor switch during the course of infection are still unclear. Several hypotheses have been proposed that may explain the late appearance of X4 viruses (38). The transmission-mutation hypothesis suggests that R5 viruses are preferentially transmitted and gradually mutate into X4 viruses, whereas the target-cell-based hypothesis emphasizes that a gradual shift in the availability of CCR5- and CXCR4-expressing cell populations is responsible for the appearance of X4 viruses. Finally, the immune system-based hypothesis suggests that X4 viruses are better recognized by the immune system and subsequently suppressed. X4 populations may emerge as a consequence of gradual immune system dysfunction.
During a study of intrapatient HIV-1 evolution, we identified several cases of recombination between coexisting R5 and X4 viruses. A hot spot for recombination was identified in the C2 region of env, and sequence analysis showed that a small part of the envelope, including the V3 region, determined coreceptor usage for both R5 and X4 recombinants. On the basis of these findings, we hypothesize that double infection followed by recombination between coexisting R5 and X4 viruses could generate less well immune system-controlled X4 variants which could be of great importance for the emergence of X4 viruses later in infection.
MATERIALS AND METHODS
Patient material.
Twenty-seven HIV-1 isolates from four patients (1865, 2239, 2242, and 2282) were selected on the basis of coreceptor usage evolution from a cohort of 53 HIV-1-infected individuals (15). The four patients were previously classified as switch virus patients (17), since viruses isolated early in infection used only CCR5, whereas the virus population isolated later in infection used both CXCR4 and CCR5 (patients 2239, 2242, and 2282) or CCR3 (patient 1865) (Table 1).
TABLE 1.
Coreceptor usage of sequential HIV-1 isolates and of V1-V3 clones from dualtropic isolates
| Patient | Time from infection (mo)a | Phenotype of isolate | No. of clones using coreceptor(s)b:
|
No. of inactive clonesc | ||
|---|---|---|---|---|---|---|
| CCR5 | CCR5 and CXCR4 | CXCR4 | ||||
| 1865 | 49 | R5 | 10 | |||
| 55 | R5 | 9 | ||||
| 61 | R3X4 | 10 | ||||
| 70 | R3X4 | 10 | ||||
| 2239 | 25 | R5 | 8 | |||
| 45 | R5 | 10 | ||||
| 68 | R5X4 | 1 | 0 | 7 | 1 | |
| 79 | R5X4 | 0 | 0 | 10 | 0 | |
| 88 | R5X4 | 4 | 0 | 4 | 2 | |
| 2242 | 18 | R5 | 10 | |||
| 45 | R5 | 8 | ||||
| 56 | R5 | 9 | ||||
| 63 | R5 | 10 | ||||
| 63 | R5 | 9 | ||||
| 64 | R5 | 10 | ||||
| 76 | R5X4 | 1 | 0 | 0 | 9 | |
| 84 | R5X4 | 7 | 0 | 1 | 2 | |
| 85:1d | R5X4 | 5 | 0 | 3 | 1 | |
| 85:2e | R5X4 | 5 | 0 | 3 | 2 | |
| 2282 | 10 | R5 | 10 | |||
| 21 | R5 | 8 | ||||
| 24 | R5 | 7 | ||||
| 41 | R5 | 10 | ||||
| 47 | R5X4 | 3 | 3 | 0 | 3 | |
| 62 | R5X4 | 3 | 5 | 0 | 1 | |
| 63 | R5X4 | 2 | 2 | 4 | 1 | |
| 70 | R5X4 | 1 | 3 | 4 | 2 | |
Time from infection was calculated as the midpoint between the last negative sample and the first positive sample.
Coreceptor usage of chimeric viruses was determined by infection of U87.CD4 cells expressing CCR5 or CXCR4.
Chimeric viruses that did not infect CCR5- or CXCR4-expressing cells.
First sample, 85 months postinfection.
Second sample, 85 months postinfection.
Generation of chimeric viruses.
Subconfluent 293T cells were transfected with 3 μg of 43XCΔV, a NheI-linearized vector containing a full-length pNL4-3 genome with the region from V1 to V3 (V1-V3) deleted (46), and with 1 μg amplified V1-V3 fragment (see below) using the calcium phosphate precipitation method. Cells were washed with phosphate-buffered saline 16 h after transfection. After 48 h, the supernatant, containing chimeric virus, was removed, cleared by centrifugation, and stored at −80°C.
Determination of coreceptor usage.
Human kidney embryonic cell line 293T cells and human glioma U87.CD4 cells, stably expressing CD4 and one of the chemokine receptors (CCR5 or CXCR4) (7) were maintained as described previously (30). Twenty-four hours prior to infection, 105 U87.CD4 cells/well were seeded in 48-well plates. For infection, 200 μl of chimeric virus was added. Cells were washed three times with Dulbecco modified Eagle medium 16 h postinfection. Six days postinfection, the cultures were analyzed for syncytium formation and p24 by using an enzyme-linked immunosorbent assay kit (Biomérieux, Boxtel, The Netherlands).
Amplification, cloning, and sequencing.
Viral RNA was extracted and purified from peripheral blood mononuclear cell culture supernatants, using Nukleospin RNA virus kit (Machery-Nagel, Germany) according to the manufacturer's instructions. Purified RNA was reverse transcribed using Superscript II (Invitrogen), and the V1-V3 region was amplified from cDNA using the Expand High Fidelity PCR system (Roche) and primers E20 and E115 (46) as described by the protocol supplied by Roche. The amplified products, approximately 900 bp (nucleotides 6002 to 6903 in HXB2; GenBank accession number AF033819), were cloned using the TOPO-TA cloning system (Invitrogen). From each isolate, 10 colonies were picked, and viral V1-V3 DNA was amplified as described above. Clones were named as follows: the patient identification number, month of isolation, and clone number (patient-month:clone number). In the case of patient 2242, two samples were taken 63 and 85 months postinfection. The second isolates for each month for this patient are designated 2242-63:2 and 2242-85:2.
Purified V1-V3 DNA was sequenced using an ABI PRISM Big Dye Termination kit (Applied Biosystems) according to the manufacturer's instructions using primers E20 (46), 793SEQ4 (5′-CAGCAGTGAGTTGATACTACTGG-3′), and JA168 and JA169 (24). Sequences were determined using ABI Prism 3100 (Applied Biosystems).
Phylogenetic analysis.
Sequences were assembled, and contigs were analyzed with CodonCode Aligner version 1.4.3 (CodonCode Corporation), aligned with ClustalX (45) and manually edited using GeneDoc. Sequences from each patient were treated as individual data sets, and Modeltest (36) was used to identify the nucleotide substitution model that fit the data best. Maximum-likelihood trees were constructed with PAUP* 4.0 (Sinauer Associates, Inc. Publishers) using heuristic searches. Statistical support of the trees was obtained by 100 bootstrap replicates using the LUNARC computer cluster (http://www.lunarc.lu.se) at Lund University, Sweden.
Recombination analysis.
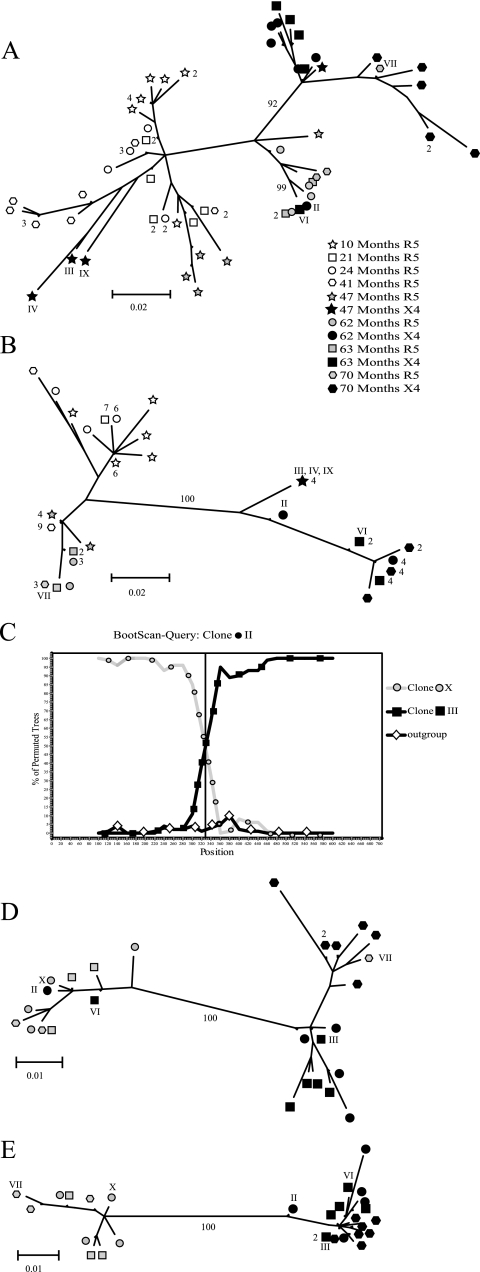
First, the data sets for each patient were split into two regions (V1/V2 and V3), and the phylogenetic trees were constructed for each data set (39). A clone was considered a recombinant if it clustered with different groups of sequences separated by significant bootstrap values (90% or more) in the two trees (see Fig. 2A and B). Putative parental sequences were identified as the sequences most similar to the recombinant in these trees. Second, we identified recombination breakpoints and parental sequences with BootScan analysis (27) using a window size of 200 bp and a 20-bp sliding step. The two putative parental sequences were considered true parental sequences if they clustered together with the recombinant in more than 90% of the permuted trees (see Fig. 2C). If both parental sequences were identified, the recombination breakpoint could be identified, the data set was split at that position, and trees were generated as described above (see Fig. 2D and E). Finally, the recombinant and parental sequences were inspected manually (see Fig. 3).
FIG. 2.
Schematic illustration of the recombination analysis, exemplified by clones from patient 2282. Phylogenetic trees were constructed from (A) the V1/V2 regions and (B) the V3 regions. Clones that clustered with different groups of sequences were considered recombinants if the groups were separated by a significant bootstrap value (≥90%) in the two trees. (C) Recombinants were analyzed by BootScan analysis for identification of the recombination breakpoints. (D and E) The data set was split at the breakpoint (nucleotide 328) and two trees were constructed to confirm the results. Different symbols represent the coreceptor usage of the clones and sampling time postinfection. Open symbols show clones from R5 isolates, gray symbols indicate phenotypically characterized R5 clones from R5X4 isolates, and black symbols represent phenotypically characterized X4 clones from R5X4 isolates. Recombinants (II, VI, and VII) and parental (III and X) clones are indicated with roman numerals. Sequences that differed by 3 nucleotides or less are represented by one terminal branch, and the number of clones that are represented at a branch is indicated. Bootstrap values that separated groups and were used for identification of recombinants are indicated.
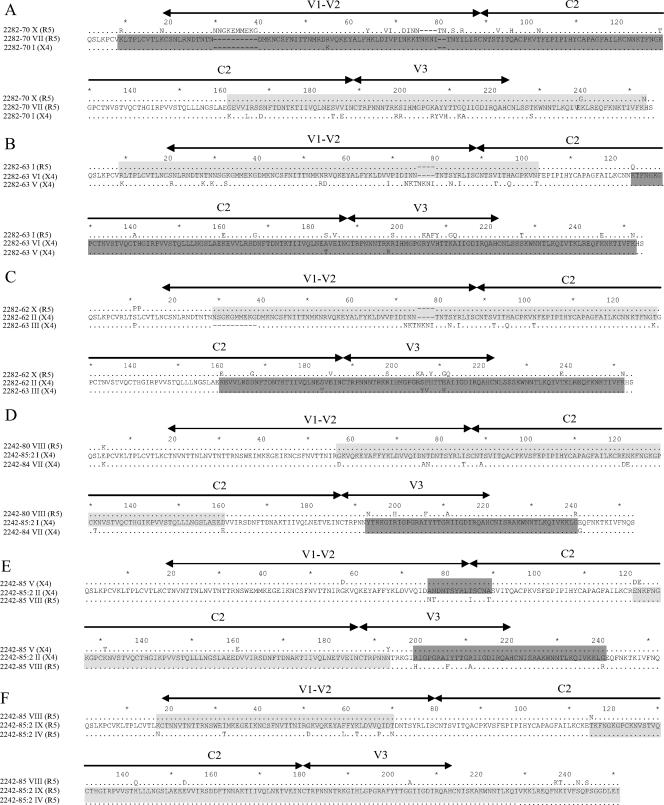
FIG. 3.
Amino acid sequences of recombinant clones. (A) R5 clone 2282-70 VII, (B) X4 clone 2282-63 VI, (C) X4 clone 2282-62 II X4, (D) X4 clone 2242-85:2 I, (E) X4 clone 2242-85:2 II, and (F) R5 clone 2242-85:2 IX. Recombinant sequences are shown in the middle of each alignment, and the parental sequences are shown above and below each recombinant sequence. Shaded regions indicate where the recombinants are most similar to one of the parental sequences. Regions shaded in light gray indicate similarity between the recombinant sequence and the R5 parental sequence, and regions highlighted in dark gray show regions of similarity between the recombinant and the X4 parent. The locations of the V1-V2, C2, and V3 regions are indicated. Dots represent identical amino acids between the recombinant and parental sequences. Recombinant clone 2242-85:2 II was most likely a result of a double-crossover event as indicated. The coreceptor usage is indicated in parentheses.
Nucleotide sequence accession numbers.
Nucleotide sequences were deposited in GenBank under the following accession numbers: DQ516085 to DQ516124 (patient 1865), DQ516125 to DQ516172 (patient 2239), DQ516173 to DQ516266 (patient 2242), and DQ516267 to DQ516338 (patient 2282).
RESULTS
Coreceptor usage.
We determined the coreceptor usage of 8 to 10 V1-V3 clones from R5X4 isolates of patients 2239, 2242, and 2282 (Table 1). Amplified V1-V3 fragments and an HIV-1 backbone were used to reconstruct chimeric viruses. The coreceptor usage of the chimeric viruses was determined by infecting cell lines expressing CD4 and either CCR5 or CXCR4. In our isolates, the R5X4 phenotype was the result of a mixture of R5 and X4 viruses in patients 2239 and 2242, whereas patient 2282 also had dualtropic R5X4 viruses (Table 1). The clones from previously characterized R5 (all patients) and R3X4 (patient 1865) isolates (16) were considered R5 and X4 clones, respectively.
Phylogenetic analysis.
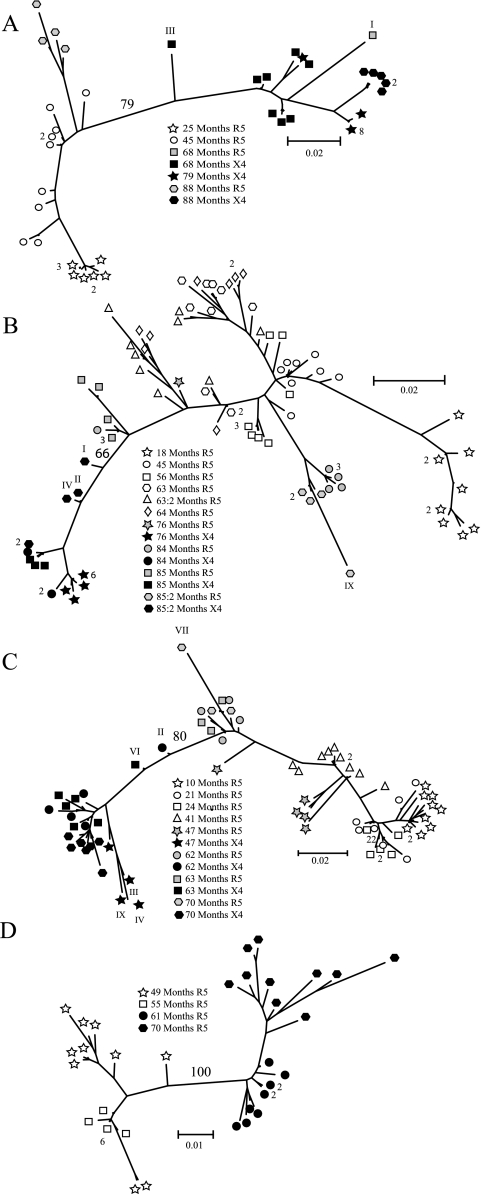
We determined the nucleotide sequences of 253 V1-V3 clones and constructed maximum-likelihood trees to study the relationships of sequences within each patient (Fig. 1A to D). In all four patients, the sequences were separated according to coreceptor usage. The bootstrap values for branches separating R5 and X4 clones were 100%, 79%, 66%, and 80% for patients 1865, 2239, 2242, and 2282, respectively. Closer inspection of the phylogenetic trees revealed several deviant sequences that either (i) were scattered between the R5 and X4 populations (2239-68 III; 2242-85:2 I, II; 2282-62 II; 2282-63 VI; Fig. 1A to C), (ii) had long branch lengths (2242-85:2 IX; 2282-47 III, IV, IX; 2282-70 VII; Fig. 1A to C), or (iii) clustered together with sequences that represented different phenotypes, i.e., one R5 clone (2239-68 I; Fig. 1A) clustered with X4 clones.
FIG. 1.
Phylogenetic relationship of HIV-1 V1-V3 clones from patients (A) 2239, (B) 2242, (C) 2282 and (D) 1865. Bootstrap values (as percentages) are indicated on branches separating R5 and X4 populations. Sequences that differed by 3 nucleotides or less are represented by one terminal branch, and the number of clones that are represented at a branch is indicated. Deviant clones (see Results) are indicated with roman numerals. Different symbols represent the coreceptor usage of the isolate and sampling time postinfection. Open symbols show clones derived from R5 isolates, gray symbols indicate phenotypically characterized R5 clones from R5X4 isolates, and black symbols represent phenotypically characterized X4 clones from R5X4 isolates or clones from R3X4 isolates.
It has been reported that intrapatient recombination occurs frequently (14, 25, 51) and that recombinant clones often deviate from other sequences in phylogenetic trees (42) in a fashion similar to our observations. We therefore anticipated that the deviant sequences represented potential recombinants, and they were subjected to further analysis.
Recombination analysis.
Since recombinants have acquired genetic material from at least two sources, they should cluster with different groups of sequences when trees are constructed from subsets of the data (39). To identify recombinants, we analyzed our data sets in three different ways. First, we generated one tree for the V1/V2 region and one for the V3 region from sequences from each patient. As exemplified by patient 2282 in Fig. 2A and B, six clones (47 III, IV, IX; 62 II; and 63 VI and 70 VII) clustered with different groups of sequences in the two trees. The cluster identity of the clones was supported by significant bootstrap values in both the V1/V2 and V3 trees, supporting that these sequences were the result of recombination events. To confirm these findings, we performed BootScan analysis. As seen in Fig. 2C, clone 62 II clustered together with clone 62 X in the 5′ end and with clone 63 III in the 3′ end. The recombination breakpoint was determined at nucleotide position 328 in the V1-V3 region analyzed (nucleotide 6473 in HXB2; GenBank accession number AF033819). Finally, to confirm the BootScan analysis, the data set was split at this position, and two trees were generated. As expected, clone 62 II clustered together with different sequences in the two trees (Fig. 2D and E).
Using this approach, we identified recombinants in all patients except for patient 1865, which was the only patient that had R5 and X4 variants that were phylogenetically separated by significant bootstrap values in the phylogenetic analysis (Fig. 1D). We identified 11 recombinants, representing 8.8% of the total 125 clones from CXCR4-using (R5X4 and R3X4) isolates (Table 1). Sequence analysis revealed that 10 of the recombinants had originated by recombination events between R5 and X4 viruses, where the majority of the recombinants used CXCR4 (8 of the 11 recombinants) (Table 2). Five of the recombinants originated from recombination between phenotypically characterized R5 and X4 clones, five from recombination between phenotypically characterized X4 clones and predicted R5 clones (on the basis of significant separation from X4 clones in V1/V2 phylogenetic tree, see Materials and Methods), and one, 2242-85:2 IX, originated from recombination event between two phenotypically characterized R5 clones (Table 2). For 6 of the 11 recombinants, both parental sequences were identified. The recombination breakpoints were located in the C2 or V3 region. For the remaining recombinant sequences, we were not able to identify the putative parental sequences (see Materials and Methods for definition of parental sequences) which corroborated identification of breakpoints.
TABLE 2.
Recombinant clones
| Patient | Clone name of recombinanta | 5′ Parental sequence nameb | 3′ Parental sequence namec | Breakpointd |
|---|---|---|---|---|
| 2239 | 68 I (R5) | 68 VII (X4) | NI (R5) | NI |
| 68 III (X4) | NI (R5) | 68 IV (X4) | NI | |
| 2242 | 85:2 I (X4) | 85 VIII (R5) | 84 VII (X4) | 6594 (C2) |
| 85:2 II (X4) | 85 VIII (R5) | 85 V (X4) | 6683 (V3) | |
| 85:2 IX (R5) | 85:2 IV (R5) | 85 VIII (R5) | 6418 (C2) | |
| 2282 | 47 III (X4) | NI (R5) | 47 V (X4) | NI |
| 47 IV (X4) | NI (R5) | 47 V (X4) | NI | |
| 47 IX (X4) | NI (R5) | 47 V (X4) | NI | |
| 62 II (X4) | 62 X (R5) | 63 III (X4) | 6473 (C2) | |
| 63 VI (X4) | 63 I (R5) | 63 V (X4) | 6499 (C2) | |
| 70 VII (R5) | 70 I (X4) | 70 X (R5) | 6531 (C2) |
Phenotypically characterized coreceptor usage is indicated in parentheses.
Phenotypically characterized coreceptor usage is indicated in parentheses. Coreceptor usage of parental sequences that were not identified (NI) was based on significant separation from X4 clones in V1/V2 phylogenetic trees.
Coreceptor usage is indicated in parentheses. The coreceptor usage of the parental sequence that was not identified (NI) was based on the coreceptor usage of the recombinant clone.
Recombination breakpoint identified using BootScan and manual inspection. The numbers are the nucleotide positions corresponding to the location in HXB2 (GenBank accession number AF033819). Envelope regions where the breakpoints were identified are indicated in parentheses. NI, not identified.
Sequence analysis.
Identification of identical parental sequences reconstituting the recombinant is expected if the recombinants were generated during PCR, since the parental sequences statistically should be in excess. In our study, the parental sequences never reconstituted the nucleotide sequence of their recombinant offspring (Fig. 3), indicating that the recombinants had accumulated additional mutations after the recombination event. In addition, three recombinants (2242-85:2 I, II, and IX) had parental sequences that belonged to phylogenetic clusters that encompassed only clones isolated from a time point that differed from the isolation time point of the recombinant (Fig. 3D to F and Table 2). These observations supported that the recombinants had originated in vivo (37).
Identification of recombination breakpoints between V1/V2 and V3 allowed us to analyze the impact on coreceptor usage of these two regions. For R5 recombinant clone 2282-70 VII, the V3 region and part of the C2 region was derived from the R5 parental sequence, whereas the remaining part of C2 and V1/V2 was contributed by the parental X4 sequence (Fig. 3A). The opposite was observed for the recombinant X4 clones 2282-63 VI (Fig. 3B) and 2282-62 II (Fig. 3C). In the case of the recombinant X4 clone 2242-85:2 I, only a small part, including V3, was derived from the X4 parental sequence, whereas the rest of the sequence was donated by the R5 parent (Fig. 3D). In fact, only five amino acids of the entire recombinant sequence (four located in V3) were X4 specific.
All recombinants had the same phenotype as the parental sequences that donated the 3′ part (including V3) of the recombinant sequences (Fig. 3 and Table 2). Our results suggest that the V1/V2 region does not impact on coreceptor usage and that the V3 region determines coreceptor usage for our recombinant clones (Fig. 3).
DISCUSSION
The study on sequence variation of the V1-V3 regions of viruses from four switch patients led to the finding that intrapatient recombination between R5 and X4 viruses occur frequently. Detection of R5/X4 recombinants is expected, since HIV-1 recombination is commonly observed, even in a single round of infection (25, 51). In 2002, Jung et al. used fluorescence in situ hybridization and found that splenocytes from two HIV-1-infected patients contained on average three or four proviruses per cell (14). It has also been shown that cells become double infected by both R5 and X4 chimeric viruses (4). These observations indicate that cells frequently become coinfected which has to occur for a recombination event to take place. Coinfection and recombination are expected to occur frequently in vivo because a substantial fraction of memory CD4+ T cells express both CCR5 and CXCR4 (2, 22).
Recombinants may be generated in vitro during the process of PCR (37) or when the virus is propagated in human peripheral blood mononuclear cells. However, several observations make in vitro recombination an unlikely explanation for the origin of the recombinants reported here. First, the nucleotide sequences of the recombinants differed from the parental sequences, that is, identical parental sequences representing the recombinant were never found. This would have been expected if the recombinants were generated in vitro (37). Second, we identified only one of the two putative parental sequences for five of the recombinants (2239-68 I and III, 2282-47 III, IV, and IX). The remaining part of the recombinant had low similarity to other clones. Third, three recombinants had one of the their parental sequences in a phylogenetic cluster that contained only clones isolated from a time point that differed from the isolation time point of the recombinant (Fig. 3 and Table 2). Taking these observations into consideration (37) and the fact that both double infection (4, 14) and intrapatient recombination are commonly observed for HIV-1 in vivo (3, 14, 19, 34, 48), we feel confident that the majority of our recombinant sequences originated in vivo (37).
Recombination events between R5 and X4 within patients have to our knowledge been reported only three times previously (3, 19, 48). Results of these studies differ from ours because none of them addressed the impact of recombination on coreceptor usage and HIV-1 pathogenesis. Here, we determined both the genotype and coreceptor usage of V1-V3 clones from sequential isolates from four switch virus patients. Characterization of the patient material in this way allowed us to couple recombination events to coreceptor usage. The majority of breakpoints that we identified were located in the C2 region (Fig. 3 and Table 2) which is in agreement with a recent report where the C2 region was identified as a hotspot for recombination (9). Identification of the recombination breakpoints together with coreceptor usage data made it possible for us to perform a detailed analysis on how the V1-V3 region impacts on coreceptor usage. We presented evidence that a small part of the envelope, including the V3 region, alone determined coreceptor specificity of the recombinant sequences studied here. Several reports have previously suggested that the V3 region is the dominant determinant for coreceptor usage (6, 8, 13, 43). It has also been suggested that other regions of env are involved in determining coreceptor usage (12, 23, 31, 32). This highlights that HIV-1 coreceptor usage, and its determinants, is complex. This is supported by a recent study which demonstrated that the V1/V2 region can compensate for loss-of-fitness mutations in the V3 region (33). A possible explanation for our results is that the clones studied here, have well-adapted, biologically optimal X4 and R5 V3 regions. Such V3 regions would be independent of the V1/V2 region in the context of coreceptor usage (33).
The appearance and dominance of X4 viruses late in infection have been debated for many years without finding a biological explanation for this phenomenon. One hypothesis addressing the coreceptor switch involves immune control (38). This hypothesis is based upon the assumption that X4 viruses are better recognized by the immune system than R5 viruses and, consequently, are suppressed. In agreement with this, in 2003, Harouse et al. showed that rhesus macaques coinfected with R5 and X4 simian-human immunodeficiency hybrid viruses showed an increase in the X4 population and a decrease in the R5 population upon depletion of CD8+ T cells (11). It has also been shown that the V1/V2 region is important for inducing neutralizing antibody response (10, 40, 44, 49, 50). A recent report also suggested that the V1/V2 region is a global regulator of the sensitivity of primary HIV-1 isolates to neutralizing antibodies (35). Furthermore, Ye et al. (50) showed that the conformational arrangement of V2 and V3 with respect to the CD4 receptor binding region of gp120 appears to be critical for the recognition by neutralizing antibodies. Thus, rearrangements in the C2 region could have a dramatic effect on the immune response directed toward the viral population. Therefore, a recombination event between an immune-resistant R5 virus and an X4 virus in the C2 region could generate variants with the potential to evade the immune response and infect cells expressing CXCR4. The broadening in cell tropism of the viral population to include CXCR4-expressing cells would result in increased CD4+ cell death and further impair the immune system, which would allow the suppressed X4 population to expand. We hypothesize that coinfection and recombination between R5 and X4 viruses may in part be responsible for the coreceptor switch late in infection.
Acknowledgments
We thank Elzbieta Vincic for expert technical assistance and Anders Kvist for valuable comments on the manuscript.
This work was supported by grants from the Swedish Research Council, the Swedish International Development Cooperation Agency/Department for Research Cooperation (SIDA/SAREC), and the Royal Physiographic Society, Lund, Sweden. DNA sequencing was performed at the SWEGENE Center of Genomic Ecology, supported by the Knut and Alice Wallenberg Foundation, Stockholm, Sweden, through the SWEGENE consortium.
Footnotes
Published ahead of print on 24 January 2007.
REFERENCES
- 1.Berger, E. A. 1997. HIV entry and tropism: the chemokine receptor connection. AIDS 11(Suppl. A):S3-S16. [PubMed] [Google Scholar]
- 2.Bleul, C. C., L. Wu, J. A. Hoxie, T. A. Springer, and C. R. Mackay. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. USA 94:1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charpentier, C., T. Nora, O. Tenaillon, F. Clavel, and A. J. Hance. 2006. Extensive recombination among human immunodeficiency virus type 1 quasispecies makes an important contribution to viral diversity in individual patients. J. Virol. 80:2472-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J., Q. Dang, D. Unutmaz, V. K. Pathak, F. Maldarelli, D. Powell, and W. S. Hu. 2005. Mechanisms of nonrandom human immunodeficiency virus type 1 infection and double infection: preference in virus entry is important but is not the sole factor. J. Virol. 79:4140-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 6.De Jong, J. J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 8.Foda, M., S. Harada, and Y. Maeda. 2001. Role of V3 independent domains on a dualtropic human immunodeficiency virus type 1 (HIV-1) envelope gp120 in CCR5 coreceptor utilization and viral infectivity. Microbiol. Immunol. 45:521-530. [DOI] [PubMed] [Google Scholar]
- 9.Galetto, R., A. Moumen, V. Giacomoni, M. Veron, P. Charneau, and M. Negroni. 2004. The structure of HIV-1 genomic RNA in the gp120 gene determines a recombination hot spot in vivo. J. Biol. Chem. 279:36625-36632. [DOI] [PubMed] [Google Scholar]
- 10.Gorny, M. K., J. P. Moore, A. J. Conley, S. Karwowska, J. Sodroski, C. Williams, S. Burda, L. J. Boots, and S. Zolla-Pazner. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J. Virol. 68:8312-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harouse, J. M., C. Buckner, A. Gettie, R. Fuller, R. Bohm, J. Blanchard, and C. Cheng-Mayer. 2003. CD8+ T cell-mediated CXC chemokine receptor 4-simian/human immunodeficiency virus suppression in dually infected rhesus macaques. Proc. Natl. Acad. Sci. USA 100:10977-10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansson, M., E. Backstrom, G. Scarlatti, A. Bjorndal, S. Matsuda, P. Rossi, J. Albert, and H. Wigzell. 2001. Length variation of glycoprotein 120 V2 region in relation to biological phenotypes and coreceptor usage of primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 17:1405-1414. [DOI] [PubMed] [Google Scholar]
- 13.Jensen, M. A., F. S. Li, A. B. van't Wout, D. C. Nickle, D. Shriner, H. X. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung, A., R. Maier, J. P. Vartanian, G. Bocharov, V. Jung, U. Fischer, E. Meese, S. Wain-Hobson, and A. Meyerhans. 2002. Multiply infected spleen cells in HIV patients. Nature 418:144. [DOI] [PubMed] [Google Scholar]
- 15.Karlsson, A., K. Parsmyr, K. Aperia, E. Sandstrom, E. M. Fenyo, and J. Albert. 1994. MT-2 cell tropism of human immunodeficiency virus type 1 isolates as a marker for response to treatment and development of drug resistance. J. Infect. Dis. 170:1367-1375. [DOI] [PubMed] [Google Scholar]
- 16.Karlsson, I., L. Antonsson, Y. Shi, A. Karlsson, J. Albert, T. Leitner, B. Olde, C. Owman, and E. M. Fenyo. 2003. HIV biological variability unveiled: frequent isolations and chimeric receptors reveal unprecedented variation of coreceptor use. AIDS 17:2561-2569. [DOI] [PubMed] [Google Scholar]
- 17.Karlsson, I., L. Antonsson, Y. Shi, M. Oberg, A. Karlsson, J. Albert, B. Olde, C. Owman, M. Jansson, and E. M. Fenyo. 2004. Coevolution of RANTES sensitivity and mode of CCR5 receptor use by human immunodeficiency virus type 1 of the R5 phenotype. J. Virol. 78:11807-11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kellam, P., and B. A. Larder. 1995. Retroviral recombination can lead to linkage of reverse transcriptase mutations that confer increased zidovudine resistance. J. Virol. 69:669-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemal, K. S., B. Foley, H. Burger, K. Anastos, H. Minkoff, C. Kitchen, S. M. Philpott, W. Gao, E. Robison, S. Holman, C. Dehner, S. Beck, W. A. Meyer III, A. Landay, A. Kovacs, J. Bremer, and B. Weiser. 2003. HIV-1 in genital tract and plasma of women: compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc. Natl. Acad. Sci. USA 100:12972-12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiwelu, I. E., I. N. Koulinska, W. M. Nkya, J. Shao, S. Kapiga, and M. Essex. 2005. Identification of CRF10_CD viruses among bar and hotel workers in Moshi, Northern Tanzania. AIDS Res. Hum. Retrovir. 21:897-900. [DOI] [PubMed] [Google Scholar]
- 21.Koulinska, I. N., T. Ndung'u, D. Mwakagile, G. Msamanga, C. Kagoma, W. Fawzi, M. Essex, and B. Renjifo. 2001. A new human immunodeficiency virus type 1 circulating recombinant form from Tanzania. AIDS Res. Hum. Retrovir. 17:423-431. [DOI] [PubMed] [Google Scholar]
- 22.Lee, B., M. Sharron, L. J. Montaner, D. Weissman, and R. W. Doms. 1999. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. USA 96:5215-5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, M. K., J. Heaton, and M. W. Cho. 1999. Identification of determinants of interaction between CXCR4 and gp120 of a dual-tropic HIV-1DH12 isolate. Virology 257:290-296. [DOI] [PubMed] [Google Scholar]
- 24.Leitner, T., G. Korovina, S. Marquina, T. Smolskaya, and J. Albert. 1996. Molecular epidemiology and MT-2 cell tropism of Russian HIV type 1 variant. AIDS Res. Hum. Retrovir. 12:1595-1603. [DOI] [PubMed] [Google Scholar]
- 25.Levy, D. N., G. M. Aldrovandi, O. Kutsch, and G. M. Shaw. 2004. Dynamics of HIV-1 recombination in its natural target cells. Proc. Natl. Acad. Sci. USA 101:4204-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, S. L., J. E. Mittler, D. C. Nickle, T. M. Mulvania, D. Shriner, A. G. Rodrigo, B. Kosloff, X. He, L. Corey, and J. I. Mullins. 2002. Selection for human immunodeficiency virus type 1 recombinants in a patient with rapid progression to AIDS. J. Virol. 76:10674-10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14(Suppl. 3):S31-S44. [PubMed] [Google Scholar]
- 30.Mild, M., A. Bjorndal, P. Medstrand, and E. M. Fenyo. 2006. Isolation of human immunodeficiency virus-type 1 (HIV-1) clones with biological and molecular properties of the primary isolate. Virology 350:58-66. [DOI] [PubMed] [Google Scholar]
- 31.Nabatov, A. A., G. Pollakis, T. Linnemann, A. Kliphius, M. I. Chalaby, and W. A. Paxton. 2004. Intrapatient alterations in the human immunodeficiency virus type 1 gp120 V1V2 and V3 regions differentially modulate coreceptor usage, virus inhibition by CC/CXC chemokines, soluble CD4, and the b12 and 2G12 monoclonal antibodies. J. Virol. 78:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogert, R. A., M. K. Lee, W. Ross, A. Buckler-White, M. A. Martin, and M. W. Cho. 2001. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolate DH12 gp120 affect coreceptor usage and cellular tropism. J. Virol. 75:5998-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pastore, C., R. Nedellec, A. Ramos, S. Pontow, L. Ratner, and D. E. Mosier. 2006. Human immunodeficiency virus type 1 coreceptor switching: V1/V2 gain-of-fitness mutations compensate for V3 loss-of-fitness mutations. J. Virol. 80:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philpott, S., H. Burger, C. Tsoukas, B. Foley, K. Anastos, C. Kitchen, and B. Weiser. 2005. Human immunodeficiency virus type 1 genomic RNA sequences in the female genital tract and blood: compartmentalization and intrapatient recombination. J. Virol. 79:353-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 37.Posada, D., K. A. Crandall, and E. C. Holmes. 2002. Recombination in evolutionary genomics. Annu. Rev. Genet. 36:75-97. [DOI] [PubMed] [Google Scholar]
- 38.Regoes, R. R., and S. Bonhoeffer. 2005. The HIV coreceptor switch: a population dynamical perspective. Trends Microbiol. 13:269-277. [DOI] [PubMed] [Google Scholar]
- 39.Salemi, M., S. L. Lamers, S. Yu, T. de Oliveira, W. M. Fitch, and M. S. McGrath. 2005. Phylodynamic analysis of human immunodeficiency virus type 1 in distinct brain compartments provides a model for the neuropathogenesis of AIDS. J. Virol. 79:11343-11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scarlatti, G., E. Tresoldi, A. Bjorndal, R. Fredriksson, C. Colognesi, H. K. Deng, M. S. Malnati, A. Plebani, A. G. Siccardi, D. R. Littman, E. M. Fenyo, and P. Lusso. 1997. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 3:1259-1265. [DOI] [PubMed] [Google Scholar]
- 42.Schierup, M. H., and J. Hein. 2000. Consequences of recombination on traditional phylogenetic analysis. Genetics 156:879-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimizu, N., Y. Haraguchi, Y. Takeuchi, Y. Soda, K. Kanbe, and H. Hoshino. 1999. Changes in and discrepancies between cell tropisms and coreceptor uses of human immunodeficiency virus type 1 induced by single point mutations at the V3 tip of the env protein. Virology 259:324-333. [DOI] [PubMed] [Google Scholar]
- 44.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trouplin, V., F. Salvatori, F. Cappello, V. Obry, A. Brelot, N. Heveker, M. Alizon, G. Scarlatti, F. Clavel, and F. Mammano. 2001. Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J. Virol. 75:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tscherning-Casper, C., G. Dolcini, P. Mauclere, E. M. Fenyo, F. Barre-Sinoussi, J. Albert, and E. Menu, for The European Network on the Study of In Utero Transmission of HIV-1. 2000. Evidence of the existence of a new circulating recombinant form of HIV type 1 subtype A/J in Cameroon. AIDS Res. Hum. Retrovir. 16:1313-1318. [DOI] [PubMed] [Google Scholar]
- 48.van Rij, R. P., M. Worobey, J. A. Visser, and H. Schuitemaker. 2003. Evolution of R5 and X4 human immunodeficiency virus type 1 gag sequences in vivo: evidence for recombination. Virology 314:451-459. [DOI] [PubMed] [Google Scholar]
- 49.Xiang, S. H., N. Doka, R. K. Choudhary, J. Sodroski, and J. E. Robinson. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retrovir. 18:1207-1217. [DOI] [PubMed] [Google Scholar]
- 50.Ye, Y., Z. H. Si, J. P. Moore, and J. Sodroski. 2000. Association of structural changes in the V2 and V3 loops of the gp120 envelope glycoprotein with acquisition of neutralization resistance in a simian-human immunodeficiency virus passaged in vivo. J. Virol. 74:11955-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhuang, J., A. E. Jetzt, G. Sun, H. Yu, G. Klarmann, Y. Ron, B. D. Preston, and J. P. Dougherty. 2002. Human immunodeficiency virus type 1 recombination: rate, fidelity, and putative hot spots. J. Virol. 76:11273-11282. [DOI] [PMC free article] [PubMed] [Google Scholar]