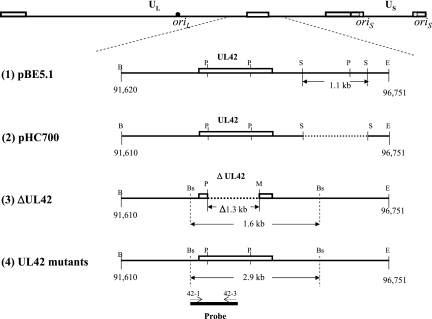
FIG. 1.
Map of UL42 plasmids and viruses. The top line shows the genomic structure of HSV-1. The relative locations of UL42 and its flanking sequences are enlarged and the region of sequences cloned and modified in plasmids (1 and 2) and recombinant viruses (3 and 4) are shown (1). pEB5.1 contains a 5.1-kbp BamHI-EcoRI DNA fragment of HSV-1 strain KOS, which includes UL42 and UL43 and partial UL41 and UL44 sequences, inserted into pGEM7zf(+) (2). pHC700 was constructed from pEB5.1 by deleting a 1.1-kbp SacII fragment (3). Recombinant virus CgalΔ42, derived from HSV-1 strain syn+, contains an inserted lacZ gene between Us9 and Us10 and a deletion of 1,258 bp from within UL42. (4). UL42 recombinants were constructed from CgalΔ42 using pHC to introduce the corresponding UL42 mutation. The probe used for Southern blotting is shown at the bottom of the figure. The open boxes above the line represent the open reading frame of UL42. Dotted lines represent deleted regions. Oligonucleotides 42-1 and 42-3 were used to amplify and label the DNA fragment. B, BamHI; Bs, BstEII; E, EcoRI; M, MluI; P, PstI; S, SacII.