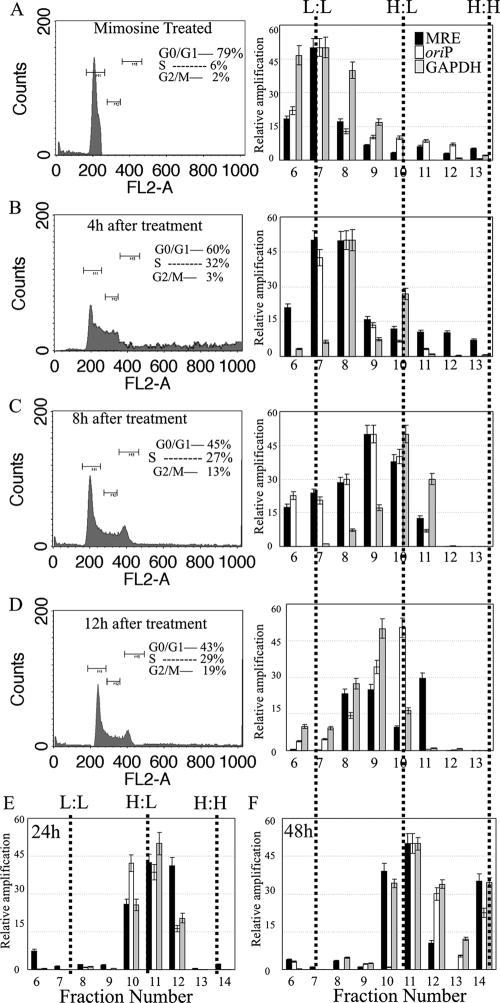
FIG. 3.
The KSHV MRE-containing plasmid replicates only once, in a cell-cycle-dependent manner. (A) 293 cells cotransfected either with pBSpuro MRE and pA3M-LANA or with pBSpuro oriP and the EBNA1 expression vector were synchronized in G1 phase by treatment with mimosine (0.5 mM) for 18 h. Synchronized cells were released into a normal medium containing 30 μg/ml BrdU. Cells were harvested at the indicated time points (0, 4, 8, 12, 24, and 48 h), and a fraction of cells was subjected to fluorescence-activated cell sorter analysis. Total DNAs from the remaining cells were extracted and sonicated to an average length of 600 bp, followed by CsCl density gradient centrifugation. Fractions of 250 μl were collected from the top, and DNAs from these fractions were then purified. Plasmid DNAs in these fractions were detected by real-time quantitative PCR using a primer targeted to the amplification of the puromycin gene on the plasmid backbone. Solid and open bars, relative amplification of the plasmid DNAs containing MRE and oriP, respectively. Quantification of cellular DNA based on the amplification of the GAPDH gene was also performed for these fractions, and the results were plotted (shaded bars) along with the plasmid DNA content in the respective fraction. The density of each fraction was detected. Dotted lines indicate fractions with L:L, H:L, and H:H densities. Only fractions 6 to 13 are shown for the 0-h (A), 4-h (B), 8-h (C), and 12-h (D) time points. Fraction 14, which is mainly the H:H fraction, is shown in the DNA from cells labeled with BrdU for 24 h (E) and 48 h (F).