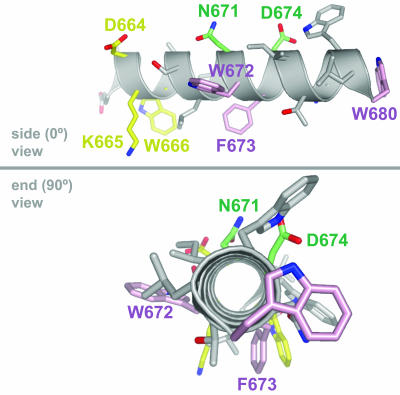
FIG. 9.
Hypothetical model of the MPER of HIV-1 gp41 as an ideal alpha helix, in side (0°) and end-on (90°) views, places 4E10 and Z13e1 crucial binding residues on opposite faces of the helix. The MPER residues that are crucial in the neutralizing activity of 2F5, Z13e1, and 4E10 are shown in yellow, green, and pink, respectively. The model of the MPER sequence DKWASLWNWFDITNWLW was created using the INSIGHT II package (Accelrys, Inc., San Diego, CA) and the PyMOL program (The PyMOL Molecular Graphics System; DeLano Scientific, San Carlos, CA) (16).