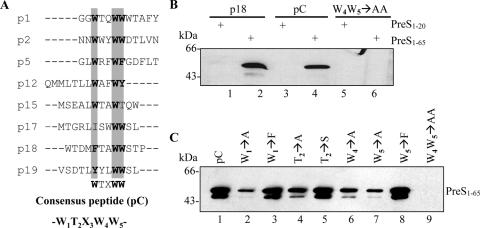
FIG. 3.
Identification and mutational analysis of a consensus PreS-binding motif. (A) Peptide alignment. Highly conserved residues are shaded and in bold. The deduced consensus sequence (pC) is W1T2X3W4W5. (B) GST pull-down assays. MBP-PreS11-65 was used for the binding assays (lanes 2, 4, and 6). Parallel controls were performed with MBP-PreS11-20 (lanes 1, 3, and 5). Lanes 1 and 2, GST-p18; lanes 3 and 4, GST-pC, with the sequence W1T2N3W4W5; lanes 5 and 6, W4W5→AA mutant of GST-pC. (C) GST pull-down assays with A, F, or S residue substitutions. The substitutions are denoted above each lane.