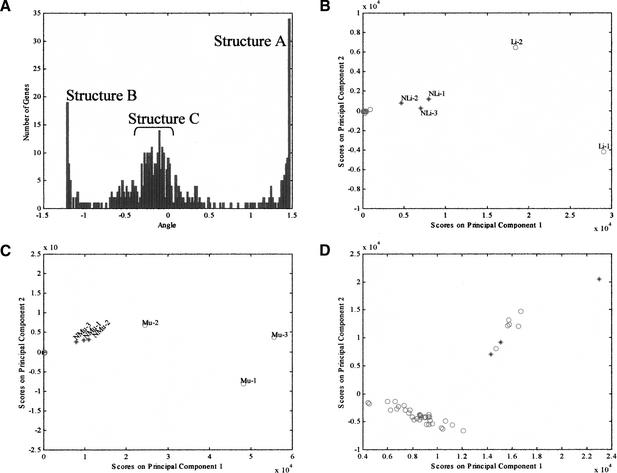
Figure 2.
Identification of tissue-specific genes and validation using new samples. (A) Histogram of the angles between the X-axis and the points defined by the two principal loadings of each gene shown in Fig. 1D. Three main features, corresponding to the linear structures shown in Fig. 1D can be discerned and are labeled as A, B, and C. (B) PCA projection of all samples using the genes in structure A. The samples in the initial data set are represented by red circles and the new samples by blue asterisks. The two liver samples in the initial data set (Li-1 and Li-2) and the new liver samples (NLi-1, NLi-2, and NLi-3) are separated from the other samples, all of which cluster at the origin. (C) Projection of all samples using the genes in structure B. The muscle samples in the initial data set (Mu-1, Mu-2, and Mu-3) are separated from the other samples along PC1. All the other tissue samples cluster at the origin. The new muscle samples are also separated when projected using these genes (NMu-1, NMu-2, and NMu-3). (D) Projection of all samples using the genes in structure C. The six brain samples in the initial data set and the three new brain samples are separated from the other samples.