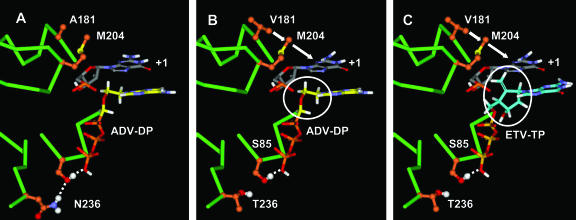
FIG. 7.
Molecular model of ADVr substitutions. (A) ADV docked into the wild-type HBV RT dNTP binding site. (B) ADV docked into the ADVr HBV RT dNTP binding site. (C) ETV docked into the ADVr HBV RT dNTP binding site. Hydrogen bonds are indicated by white dotted lines. The model highlights the preorganized nature of the wild-type dNTP binding site. The ADVr mutations at N236 and A181 result in a loss of secondary stabilization. The circled moieties in panels B and C are the deoxyribose mimics in ADV and ETV, respectively. Note that the binding affinity of the flexible ADV is predictably reduced for the slightly less preorganized ADVr dNTP binding site, while binding of more rigid NRTIs, such as ETV and LVD, is not.