Figure 7.
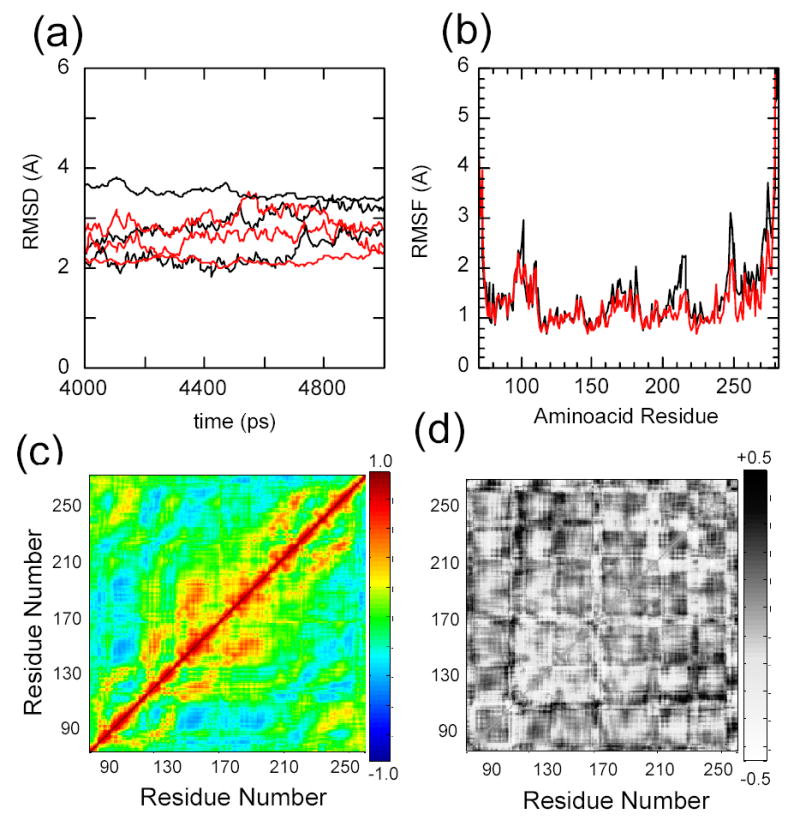
Molecular dynamics simulations of IκBα with the initial coordinates taken from the co-crystal structure of IκBα in complex with NF-κB (pdb accession 1NFI, chain E), in explicit solvent, relaxed for 5 nsec (black). The C186P·A220P mutations were computationally introduced prior to the relaxation (red). Three independent trajectories were run for each protein a) Root mean square deviation of the proteins relative to the co-crystal form. b) Average root mean square fluctuations of the individual amino acids. c) Average Cα covariance matrix of wild type IκBα. d) Cα covariance difference between wild type IκBα and the C186P·A220P mutant.
