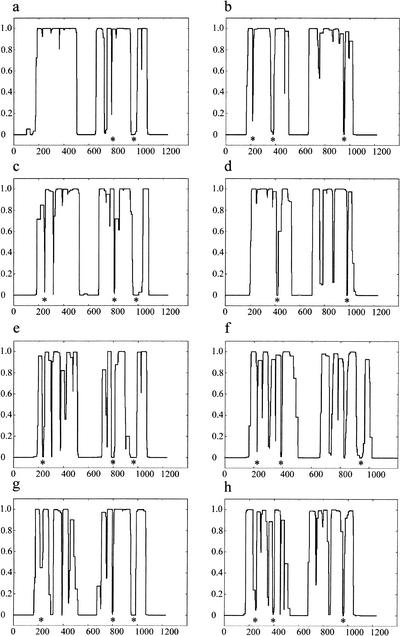
Figure 1.
COILS prediction of SMC1 and SMC3 orthologs. The COILS output for Homo sapiens SMC1 (a), H. sapiens SMC3 (b) , Drosophila melanogaster SMC1 (c), D. melanogaster SMC3 (d), fission yeast (Schizosaccharomyces pombe) SMC1 (e), fission yeast SMC3 (f), budding yeast (Saccharomyces cerevisiae) SMC1 (g), and budding yeast SMC3 (h) . The vertical axis denotes the probability of the amino acid sequence adopting a coiled-coil structure based on a scanning window of 28 residues. Although the original COILS output included windows of 14 and 21 residues, these have not been displayed for clarity. Conserved disruptions in the coiled-coil structure prediction to the cohesin SMC sequences are indicated beneath the COILS profile with an asterisk.