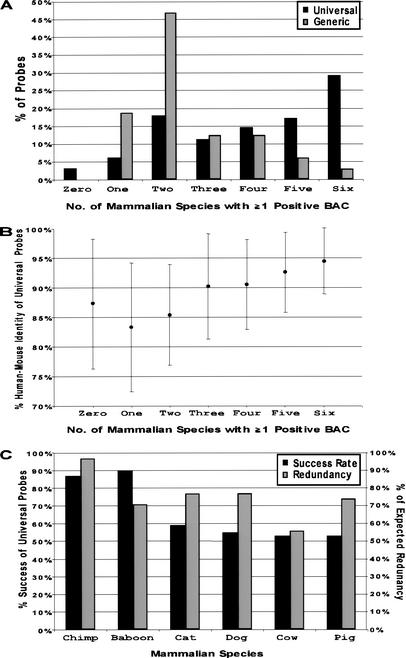
Figure 2.
Performance of universal overgo hybridization probes designed from human-mouse sequence alignments. A set of universal overgo hybridization probes (n = 341) was designed from orthologous human-mouse sequence alignments and used to screen six mammalian bacterial artificial chromosome (BAC) libraries. (A) The percent of the universal probes that yielded at least one positive BAC from the indicated numbers of mammalian species is indicated. In parallel, a set of generic overgo hybridization probes (n = 32) was designed from single-copy human sequences (not from human-mouse sequence alignments). The analogous performance of the generic probes for screening the same set of mammalian BAC libraries also is shown. (B) The above universal probes were grouped based on their relative performance in screening the six mammalian BAC libraries (i.e., the number of mammalian species for which at least one positive BAC was identified). The average percent human-mouse sequence identity was calculated for each group, with the indicated error bars reflecting one standard deviation from the mean. (C) The percent of the above universal probes that identified at least one positive BAC for each mammalian species is indicated. Also shown is the percent of the expected clone redundancy provided by the isolated BACs for each mammalian species, which reflects the ratio of the average redundancy obtained with the set of universal probes to the expected redundancy (as calculated for each BAC library).