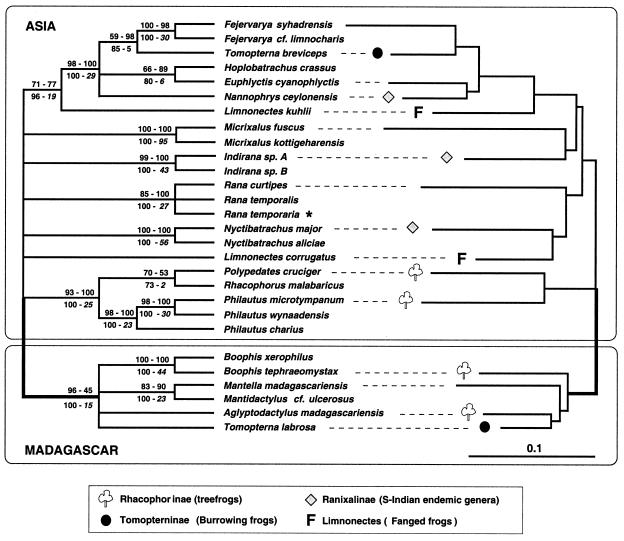
Figure 1.
Unrooted molecular phylogeny of Asian and Madagascan Ranidae. MP analyses of the mtDNA and nuDNA data sets both yield majority rule bootstrap consensus trees compatible with that shown on the Left. The first and second values given above the branches are bootstrap values (1,000 replicates, 50 random additions each) for the analyses of mtDNA and nuDNA data sets, respectively. The first and second values below the branches indicate bootstrap values and Bremer support, respectively, for the analysis of the combined mtDNA and nuDNA data sets. Right shows the best tree under ML. Wherever the tree is rooted, there is no possibility to make Rhacophorinae, Tomopterninae, Ranixalinae, or the genus Limnonectes monophyletic. This finding indicates multiple convergences of ecomorphs among and within Madagascar, the Indian subcontinent, and Asia. Monophyly of Madagascan Ranidae (bold branch) is strongly supported (see text for details). We included a European Rana species (indicated by an asterisk) which, as expected, clusters with Asian members of the genus.