Figure 1.
Comparative Analysis of AOX Expression in the Wild Type and CMSII Mutant.
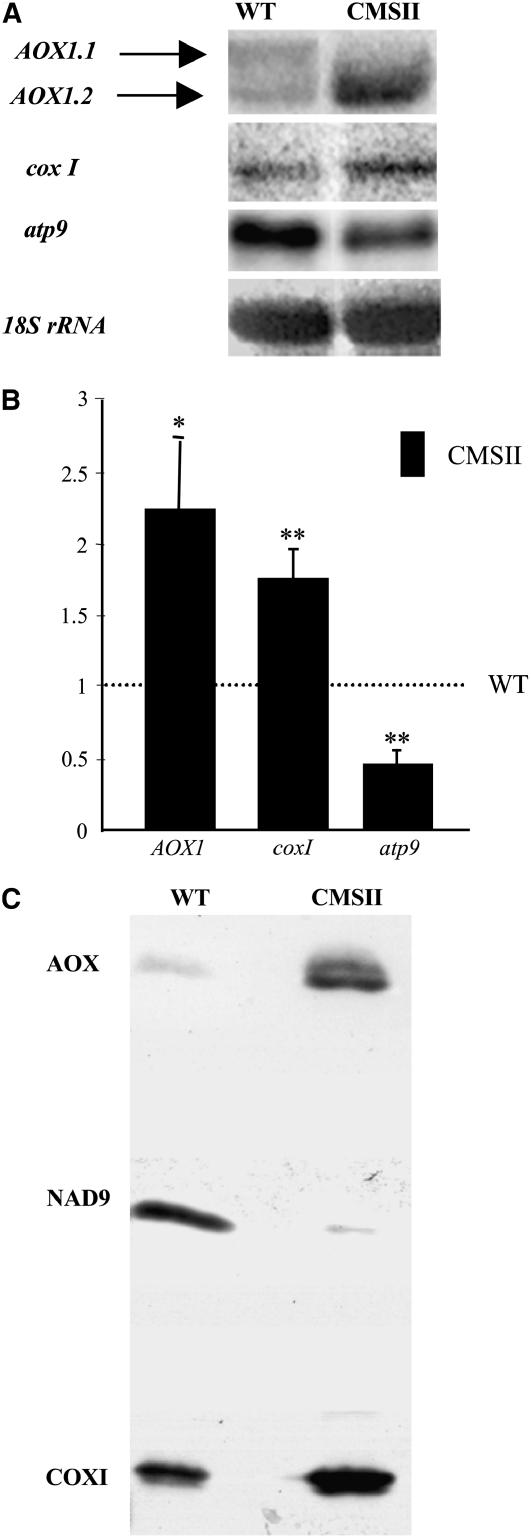
(A) Total RNA was extracted from mature leaves of wild-type and CMSII plants and subjected to RNA gel blot analysis (12 μg RNA per lane) using as probes an N. sylvestris AOX1 internal cDNA fragment (Sabar et al., 2000) and atp9 and coxI mitochondrial sequences (Brangeon et al., 2000); 18S rDNA was used as a loading control.
(B) Relative abundance of AOX1 (pooled signals of AOX1.1 and AOX1.2), coxI, and atp9 transcripts in CMSII compared with the wild type (fixed to 1, dashed line). RNA gel blot signals were scanned with Scion imaging software and expressed as the values of the ratio: integrated density of the signal/integrated density of the 18S rRNA signal. Values are the means ± se from seven independent experiments. AOX1 and coxI transcripts are more abundant, and atp9 transcripts are less abundant in CMSII than in the wild type (*P < 0.05; **P < 0.01).
(C) Immunoblot of mitochondrial proteins from wild-type and CMSII leaves using a monoclonal Sauromatum guttatum anti-AOX antibody. Antibodies against NAD9, which is greatly decreased in CMSII leaves as a result of complex I assembly failure (Gutierres et al., 1997), and COXI were used as controls for loading and sample identity on the same membrane. Mitochondria were isolated from leaves as in Michalecka et al. (2004). Proteins (20 μg), as determined by the bicinchoninic acid method (Sigma-Aldrich), were loaded per lane by reducing SDS-PAGE on 10% acrylamide gels and wet electroblotted to nitrocellulose membranes in 10 mM CAPS, pH 11, and 20% methanol (v/v).