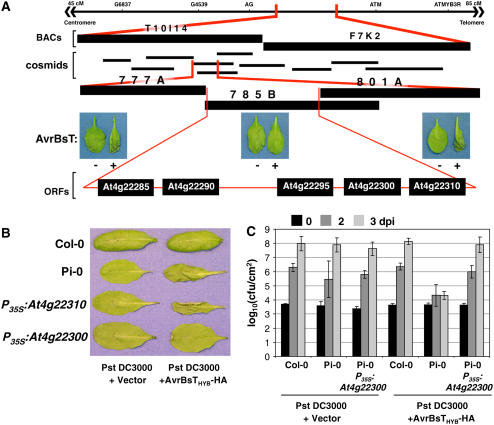
Figure 5.
Cloning of SOBER1.
(A) Physical mapping of the SOBER1 locus. The top line represents the genomic region on chromosome IV between markers AG and ATM, to which the SOBER1 locus was first mapped. Two Col-0 BACs (T10I14 and F7K2) and 10 cosmid clones spanning this region were then isolated. Col-0 cosmid clones 777A, 785B, and 801A were independently transformed into Pi-0 for complementation analysis. T1 transgenic lines were infected with a 3 × 108 cfu/mL suspension of Pst DC3000 pVSP61 and Pst DC3000 pVSP61(avrRpt21-100-avrBsT11-350-HA), designated as AvrBsT − and +, respectively, and scored for HR at 10 to 12 h after inoculation. The complementing Col-0 cosmid clone 785B contains five genes: At4g22285, At4g22290, At4g22295, At4g22300, and At4g22310.
(B) Functional complementation of sober1-1 with At4g22300. Col-0, Pi-0, and T1 Pi-0 transgenic lines expressing P35S-At4g22310 and P35S-At4g22300 were infected with a 3 × 108 cfu/mL suspension of Pst DC3000 pVSP61 and Pst DC3000 pVSP61(avrRpt21-100-avrBsT11-350-HA), designated as Pst DC3000 and Pst DC3000 + AvrBsTHyb-HA, respectively, and scored for HR at 10 to 12 h after inoculation.
(C) In planta bacterial growth in a T1 Pi-0 transgenic line expressing P35S-At4g22300. Leaves were hand-inoculated with the 105 cfu/mL suspension of bacteria described for (B). Bacteria present in leaves were monitored at 0 d (black bars), 2 d (dark gray bars), and 3 d (light gray bars) after inoculation (dpi). Data points represent the mean log10 (cfu/cm) ± sample sd. The data represent phenotypes observed in at least three independent T1 Pi-0 transgenic lines.