Abstract
Arabidopsis thaliana HYPONASTIC LEAVES1 (HYL1) is a microRNA (miRNA) biogenesis protein that contains two N-terminal double-stranded RNA binding domains (dsRBDs), a putative nuclear localization site (NLS), and a putative protein–protein interaction domain. The interaction of HYL1 with DICER-LIKE1 is important for the efficient and precise processing of miRNA primary transcripts in plant miRNA biogenesis. To define the roles of the various domains of HYL1 in miRNA processing and the miRNA-directed phenotype, we transferred a series of HYL1 deletion constructs into hyl1 null mutants. The N-terminal region containing dsRBD1 and dsRBD2 completely rescued the mutant phenotype of hyl1, triggering the accumulation of miR166 and miR160 and resulting in reduced mRNA levels of the targeted genes. In vivo biochemical analysis of the HYL1-containing complexes from the transgenic plants revealed that the N-terminal dsRBDs of HYL1 were sufficient for processing miRNA precursors and the generation of mature miRNA. Transient and stable expression analysis demonstrated that the putative NLS domain was indeed the nuclear localization signal, whereas the N-terminal region containing the dsRBDs was not restricted to the nucleus. We suggest that the N-terminal dsRBDs fulfill the function of the whole HYL1 and thus play an essential role in miRNA processing and miRNA-directed silencing of targeted genes.
INTRODUCTION
MicroRNAs (miRNAs) are regulatory RNAs of ∼22 nucleotides that derive from stem-loop regions of endogenous precursor transcripts (Ambros, 2004; Bartel, 2004). Many miRNAs are known to pair to the complementary regions of target mRNAs to mediate posttranscriptional repression. In animals, miRNA primary transcripts (pri-miRNAs) are trimmed in the nucleus into miRNA precursors (pre-miRNAs) by an RNase III–like enzyme called Drosha (Lee et al., 2003). The pre-miRNAs are exported to the cytoplasm by exportin-5 (Yi et al., 2003; Lund et al., 2004) and are cleaved there to generate mature miRNAs by another RNase III–like enzyme called Dicer (Bernstein et al., 2001). These miRNAs are then incorporated into the RNA-induced silencing complex (RISC) endonuclease and negatively regulate the genes controlling several developmental processes by degradation of the mRNAs they pair with. In plants, miRNA biogenesis and functional pathways have been partly described. miRNAs are processed from hairpin precursor RNAs by an RNase III endonuclease known as DICER-LIKE1 (DCL1) (Park et al., 2002; Reinhart et al., 2002). Posttranscriptional repression occurs primarily through miRNA-directed cleavage of target mRNAs (Llave et al., 2002; Tang et al., 2003; Schwab et al., 2005). Proper miRNA accumulation depends on the activity of the nuclear proteins DCL1 and HUA ENHANCER1 (HEN1) (Park et al., 2002; Reinhart et al., 2002; Boutet et al., 2003). DCL1 is thought to catalyze the processing of pri-miRNAs and pre-miRNAs, subsequently producing a wide variety of miRNAs that control the expression of various important genes, such as transcription factors and development-related genes (Park et al., 2002; Reinhart et al., 2002; Bartel and Bartel, 2003; Papp et al., 2003). Genetic evidence suggests that the double-stranded RNA (dsRNA) binding domain (dsRBD) of DCL1 is essential for its in vivo function (Jacobsen et al., 1999; Schauer et al., 2002).
HYL1 is a member of a family of miRNA biogenesis proteins and participates in miRNA accumulation in coordination with DCL1 and HEN1. The dcl1 null alleles are embryo-lethal (Schauer et al., 2002), whereas hyl1 null alleles exhibit reduced miRNA levels and developmental defects that overlap with those of partial loss-of-function dcl1 mutants. Recently, the role played by HYL1 was identified in miRNA-mediated gene regulation: HYL1 has a similar function to those of DCL1 and HEN1 (Han et al., 2004; Vaucheret et al., 2004; Vazquez et al., 2004a, 2004b). Our previous study showed that HYL1 maintains adaxial/abaxial polarity of growing leaves by altering the level of miR165/166, which directs the cleavage of REVOLUTA (REV) transcripts (Yu et al., 2005).
The HYL1 protein has two dsRBDs (dsRBD1 and dsRBD2) in its N-terminal half, and it preferentially binds dsRNA in vitro (Lu and Fedoroff, 2000). HYL1 also has a putative nuclear localization site (NLS) and a putative protein–protein interaction (PPI) domain. Although HYL1 has been postulated to be located in the nucleus, the function of the NLS in the localization of HYL1 for miRNA biogenesis has not been identified. Thus, evidence is necessary to support the prediction of this putative domain. RNA binding proteins play important roles in a wide variety of cellular and developmental functions ranging from RNA processing and editing to RNA transport, localization, stabilization, and translational control of certain mRNAs (Bandziulius et al., 1989). The conserved N-terminal dsRBDs identified in these dsRNA binding proteins consist of ∼70 amino acid residues that form an α-β-β-β-α fold, two a-helices of which interface with the bound dsRNA molecule (St. Johnston et al., 1992; Ryter and Schultz, 1998). In recent years, the functions of dsRNA binding proteins in the developmental regulation of plants have been exploited intensively (Bartel and Bartel, 2003).
Biochemical deletion analysis indicates that HYL1 interacts strongly with DCL1. The dsRBD1 of HYL1 is essential for dsRNA binding in vitro, whereas dsRBD2 contributes to its PPI activity (Hiraguri et al., 2005). The interaction between DCL1 and HYL1 is important for the efficient and precise processing of pri-miRNA during plant miRNA biogenesis (Kurihara et al., 2006). It remains controversial whether or not the putative NLS and PPI domains of HYL1 serve as important components for miRNA biosynthesis and regulatory functions during development. As the NLS and PPI domains of HYL1 are predicted to be a nuclear localization signal and a PPI site, respectively, the question remains whether HYL1 without NLS and PPI domains would be able to function in the production of miRNAs and in the establishment of various developmental processes.
The hyl1 mutation causes a series of phenotypic abnormalities, such as leaf hyponasty, delayed flowering, altered root gravitropic response, and altered responses to several hormones (Lu and Fedoroff, 2000). A recent study showed that the hyl1 mutant of Arabidopsis thaliana is characterized by a transverse curvature of leaves and that the adaxial/abaxial polarity of growing leaves mediated by miR165/166 is perturbed in hyl1 mutants (Yu et al., 2005). Meanwhile, one report mentions that the amounts of eight miRNAs, including miR160, are reduced and that the transcripts of AUXIN RESPONSE FACTOR17 (ARF17) targeted by miR160 are increased in hyl1 mutants (Vazquez et al., 2004a). It is believed that miRNA-mediated gene regulation and the endogenous small interfering RNA pathway play an important role in leaf polarity and the auxin response.
In this study, we made several HYL1 mutant constructs and transformed them into hyl1 plants. Using the resulting transgenic plants, we determined the phenotypic consequences of exogenous HYL1 domains in hyl1 mutants. The hyl1 plants transformed with a HYL1 cDNA containing only the two N-terminal dsRBDs displayed complete rescue of the phenotypes conferred by the hyl1 mutant. This finding indicates that the two dsRBDs of HYL1 are sufficient to rescue the mutant phenotypes and that the C-terminal NLS and PPI domains are not required. The N-terminal dsRBDs are the most important domains in HYL1, being required for pre-miRNA processing in plants. Comparison of the transgenic plants expressing the HYL1 deletion mutant constructs and wild-type plants revealed that the N-terminal dsRBDs of HYL1 restore the accumulation of miR165, miR160, and miR319 and guide the posttranscriptional silencing of REV and ARF17 genes, suggesting that the N-terminal dsRBDs fulfill the function of HYL1 and play an essential role in leaf development.
RESULTS
The N-Terminal dsRBDs of HYL1 Are Sufficient to Rescue the Phenotype Conferred by hyl1
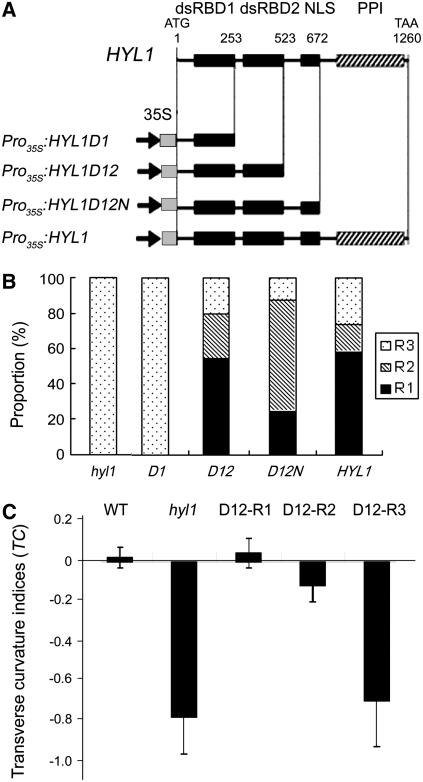
hyl1 mutant plants with HYL1 disrupted in the second exon by a Dissociation (Ds) insertion displayed the striking phenotype of incurved (hyponastic) leaves (Lu and Fedoroff, 2000; Yu et al., 2005). To determine whether the featured domains of HYL1 rescue the phenotype conferred by hyl1, we prepared plant gene expression constructs of a series of HYL1 deletion mutants (Figure 1A). HYL1D1 (HYL1 lacking its dsRBD2, NLS, and PPI) consists of a short N-terminal region and dsRBD1 fragment; HYL1D12 (HYL1 lacking its NLS and PPI) consists of an N-terminal region containing dsRBD1 and dsRBD2; and HYL1D12N (HYL1 lacking its PPI) consists of an N-terminal region and the fragment containing dsRBD1, dsRBD2, and NLS. These sequences were inserted into binary vectors under the control of the 35S promoter of the Cauliflower mosaic virus and transformed back into the hyl1 mutant plants. The resulting transgenic plants were identified individually by PCR and DNA gel blot hybridization.
Figure 1.
HYL1 Constructs and Phenotypes of the Transgenic Plants.
(A) Diagrams of the deletion mutants of the HYL1 gene and the featured domains of the HYL1 protein. The locations of the four conserved domains (dsRBD1, dsRBD2, NLS, and PPI) are indicated by squares (at top). The positions of the first and last nucleotides in the deletion mutants are numbered from the initial ATG (above HYL1 protein diagram). The constructs Pro35S:HYL1D1, Pro35S:HYL1D12, Pro35S:HYL1D12N, and Pro35S:HYL1 are shown.
(B) Proportion of rescued plants within the T1 population. D1, D12, D12N, and HYL1 are four T1 populations of hyl1 plants transformed with Pro35S:HYL1D1, Pro35S:HYL1D12, Pro35S:HYL1D12N, and Pro35S:HYL1, respectively. R1 indicates plants that were completely rescued, R2 indicates plants that were strongly rescued, and R3 indicates weakly rescued plants. The number of transformants analyzed was 28 to 58 for each segregating population.
(C) Transverse curvature indices of the rescued plants. The fourth leaves of 25-d-old plants were cut transversely at the widest extreme and pressed on the desk for measurement of pressed width (pw) when the distance between the lateral margins (lm) of the leaves at the widest extreme was measured. The transverse curvature index (TC) is calculated with the formulae TCu = (lm − pw)/pw and TCd = (pw − lm)/pw. Error bars represent the sd of >20 seedlings.
To quantify the rescue of the phenotype conferred by hyl1 caused by the various deletion mutants of HYL1, we formulated a transverse curvature index (TC) of leaves. For quantitative measurement of leaf incurvature (upward curvature) (u), the transverse curvature index is defined as TCu = (lm − pw)/pw, where lm is the distance between the lateral margins of incurved leaves and pw denotes pressed leaf width. For leaf downward curvature (d), the transverse curvature index is defined as TCd = (pw − lm)/pw. With the leaf curvature index as a quantitative measurement of allelic strength, the transgenic plants were classified into three classes: R1, completely rescued plants with flat leaves (TC = −0.10 ∼+0.10); R2, partially rescued plants with weakly curved leaves (TC = −0.11∼−0.30); and R3, nonrescued plants with strongly curved leaves (TC = −0.31∼−0.90). Within the T1 population, 58% of Pro35S:HYL1D12, 25% of Pro35S:HYL1D12N, and 53% of Pro35S:HYL1 plants belonged to the R1 class, whereas 30% of Pro35S:HYL1D12, 66% of Pro35S:HYL1D12N, and 19% of Pro35S:HYL1 plants belonged to the R2 class (Figures 1B and 2A; see Supplemental Table 1 online). By contrast, all Pro35S:HYL1D1 plants belonged to the R3 class. The proportion of rescued plants among the Pro35S:HYL1D12 population was 85%, approximately equal to that of the Pro35S:HYL1 population, meaning that Pro35S:HYL1D12 rescued at the same efficiency as Pro35S:HYL1.
Figure 2.
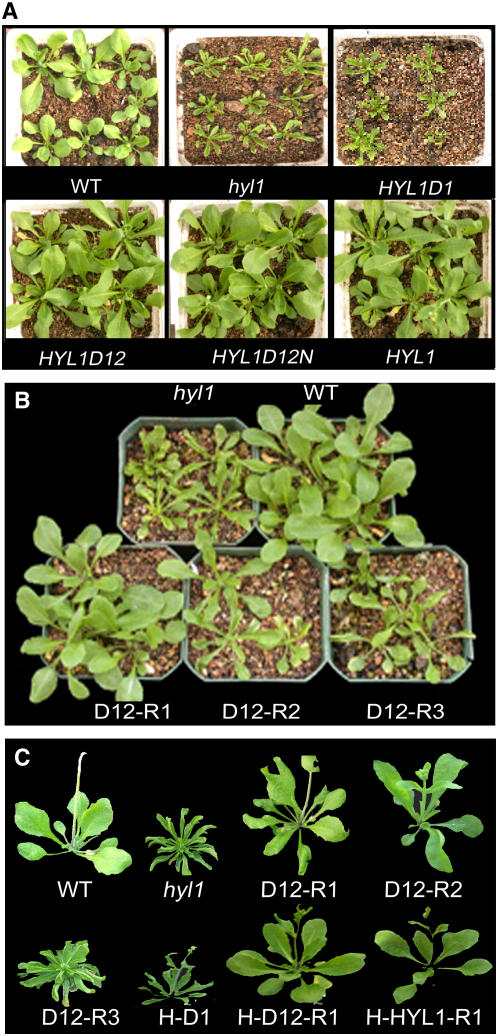
Phenotypes of the Rescued and Nonrescued Plants.
(A) T1 plants (21-d-old) of hyl1 mutants transformed with four deletion mutants of the HYL1 gene under the control of the 35S promoter. D1, D12, D12N, and HYL1 are four T1 populations of hyl1 plants transformed with Pro35S:HYL1D1, Pro35S:HYL1D12, Pro35S:HYL1D12N, and Pro35S:HYL1, respectively. Most Pro35S:HYL1D12 (D12) and Pro35S:HYL1D12N (D12N) plants resemble wild-type plants, showing flat leaves, except that these are slightly larger than those of wild-type plants. Pro35S:HYL1D1 (D1) plants look like hyl1 mutants, with incurved leaves.
(B) Shoot phenotypes of D12 lines (21-d-old) homozygous for Pro35S:HYL1D12 in the T3 generation. D12-R1, D12-R2, and D12-R3 are three classes of rescued plants. Comparison is made with hyl1 and wild-type shoots.
(C) Individual bolting plants (30-d-old) rescued from hyl1 mutants. D12-R1, D12-R2, and D12-R3 are three classes of rescued plants expressing Pro35S:HYL1D12 under the control of the cauliflower mosaic virus 35S promoter. H-D1, H-D12-R1, and H-HYL1-R1 represent transgenic lines expressing ProHYL1:HYL1D1, ProHYL1:HYL1D12, and ProHYL1:HYL1, respectively, under the control of the native promoter of the HYL1 gene.
For phenotypic comparison, we isolated transgenic lines homozygous for Pro35S:HYL1D12, Pro35S:HYL1D12N, and Pro35S:HYL1. According to the TC values, 18 independent D12-R1 lines expressing Pro35S:HYL1D12 and 25 D12N-R1 lines expressing Pro35S:HYL1D12N were defined, all of which contained BAR sequences, indicating that they were not wild-type contaminants. These plants were self-pollinated to generate T3 populations and subsequently confirmed to be homozygous for the corresponding transgenes either by their segregation or by PCR. As indicated in Figure 1C, the TC values of D12-R1 were >0 and almost the same as those of wild-type plants, the leaves of which curved downward slightly. Meanwhile, the TC values of D12-R3 were slightly greater than those of hyl1 mutants. No significant difference in genetics or morphometric behavior was recognized between D12-R1, D12N-R1, and HYL1-R1 plants; therefore, only the D12-R1 line is described below, unless indicated otherwise. Like HYL1-R1, D12-R1 and D12N-R1 seedlings displayed flat leaves similar to those of wild-type plants, indicating that Pro35S:HYL1D12 and Pro35S:HYL1D12N rescued the phenotype conferred by hyl1 completely in these seedlings (Figures 2B and 2C). By contrast, all D1 seedlings remained as deficient in leaf flatness as hyl1 mutants. They looked like hyl1 mutants and had a series of phenotypic abnormalities, such as small stature and delayed flowering. Thus, it seems that Pro35S:HYL1D1 does not rescue the hyl1 mutant phenotype. The rosette leaves of D12-R1, D12-R2, and D12-R3 plants at bolting stages displayed a similar rescue to those of plants at the seedling stage (Figure 2C). On the other hand, Pro35S:HYL1D2N (HYL1 lacking its N-terminal dsRBD1 and PPI) and Pro35S:HYL1D2NP (HYL1 lacking its N-terminal dsRBD1) showed no obvious rescue of the phenotype conferred by hyl1 (see Supplemental Table 1 online). These findings suggest that both dsRBD1 and dsRBD2 are essential for the role of HYL1 in the complete rescue of the phenotype conferred by hyl1.
In the transgenic plants, the protein fragments that are expressed at very high levels under the strong 35S promoter may take on functions normally not performed under physiological concentrations of the protein. To exclude this possibility, we used a native promoter of the HYL1 gene to drive the expression of the deletion mutants by construction of ProHYL1:HYL1D1, ProHYL1:HYL1D12, and ProHYL1:HYL1 constructs (see Supplemental Figure 1 online). Within the T1 population of ProHYL1:HYL1D12, 64% of plants were rescued completely and 25% were rescued partially, at the same efficiency as Pro35S:HYL1D12. Like D12-R1 plants with the 35S promoter, the H-D12 (ProHYL1:HYL1D12) plants and H-D12N-R1 (ProHYL1:HYL1D12N) plants with the native promoter displayed flat leaves, suggesting that ProHYL1:HYL1D12 and ProHYL1:HYL1D12N rescue the phenotype conferred by hyl1 under the control of the native promoter (Figure 2C). Indeed, the native promoter of HYL1 plays the same role as the 35S promoter in the rescue of the phenotype conferred by hyl1. We deduce that the N-terminal region with two N-terminal dsRBDs is sufficient for the role of HYL1 in the complete rescue of the hyl1 mutant phenotype.
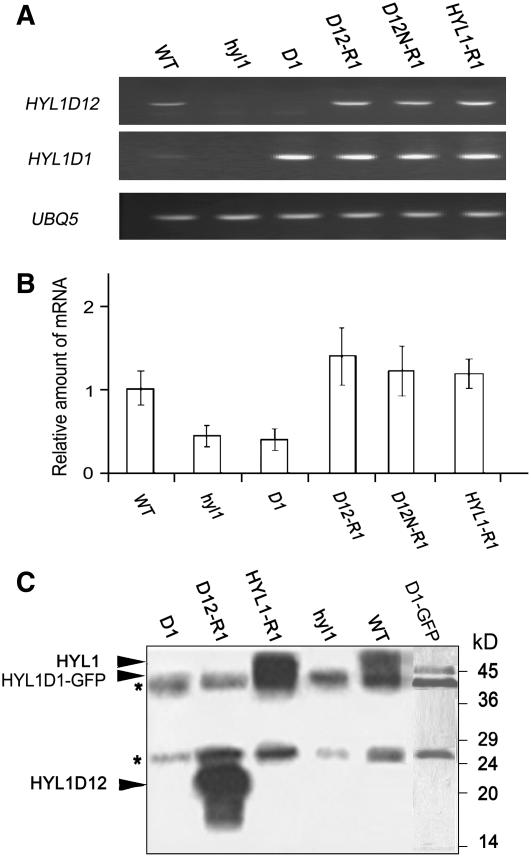
To identify the deleted versions of HYL1 in transgenic plants, we performed PCR of genomic DNA using various pairs of primers. When a pair of primers (E-HYL1-5 and E-HYL1D1-3) (see Supplemental Table 2 online) spanning the first N-terminal dsRBD was used, PCR products were observed in all plant lines examined except hyl1 mutants (Figure 3A); when a pair of primers (E-HYL1-5 and E-HYL1D12-3) spanning the N-terminal region with two dsRBDs was selected, the PCR products were observed in all plant lines examined except hyl1 mutants and D1 plants. Although the first pair of primers generated the PCR products in D1 and H-D1 plants, the rescue of the hyl1 mutant phenotype was not recognized in these plants. The presence of exogenous cDNA fragments indicates that each deletion mutant of the HYL1 gene was integrated into the genomes of transgenic plants, as expected.
Figure 3.
Expression of HYL1 Genes and Protein Gel Blot of HYL1.
(A) Detection of the transgenes. Primers specific for the HYL1D1 and HYL1D12 regions were adopted. The presence of the deletion mutants of HYL1 was measured by PCR.
(B) Expression of the HYL1 gene. Plants were transformed with either an empty vector or Pro35S:HYL1 constructs. Primers specific for HYL1D12 were adopted. The expression level of HYL1 transcripts was measured by real-time quantitative PCR in each indicated line and normalized to the expression level in wild-type plants. Error bars represent sd of three biological replicates.
(C) Protein gel blot analysis to detect the mutant protein variants of HYL1. Arrowheads point to the bands containing HYL1 and HYL1D12. The molecular masses of marker proteins are indicated at right. Asterisks denote contaminating polypeptides that were also present in the control immunoprecipitates from hyl1 extract. D1, D12-R1, D12N-R1, and HYL1-R1 are the transgenic lines expressing Pro35S:HYL1D1, Pro35S:HYL1D12, Pro35S:HYL1D12N, and Pro35S:HYL1, respectively. hyl1 mutants and wild-type plants were used as the negative and positive controls, respectively.
To address the involvement of the HYL1 deletion mutants in the differential rescue of the phenotypes conferred by hyl1, real-time quantitative PCR was performed to analyze the expression patterns of exogenous HYL1 deletion mutants in transgenic plants. To compare the expression levels of HYL1 among the transgenic lines with HYL1 deletion mutants, HYL1 transcripts in D12-R1 and D12N-R1 plants were assayed. As indicated in Figure 3B, the expression levels of HYL1 in D12-R1 and D12N-R1 plants were much greater than those of wild-type plants, even though the leaves of the former were as flat as those of the latter.
The rescue of the phenotype conferred by hyl1 by HYL1 deletion mutants depends on the functions of the mutant proteins. To address the existence and stability of the mutant protein variants of HYL1 in transgenic plants, we performed protein gel blot analysis. The mutant protein variant, HYL1D12, was found in D12-R1 and D12N-R1 seedlings, whereas HYL1D1 was not detectable in D1 plants (Figure 3C). Unlike HYL1D1, HYL1D1-GFP (for green fluorescent protein) was detected by protein gel blot in D1-GFP plants expressing Pro35S:HYL1D1-GFP, indicating that HYL1D1 was present but not detectable in itself. It is possible that the band of such a small molecule was too faint to observe under our detection conditions. Regarding HYL1D12, the H-HYL1D12 protein variant was also recognized in H-D12-R1 (ProHYL1:HYL1D12) plants, which contain the native promoter of HYL1. Nevertheless, the amounts of these proteins, which were expressed in transgenic plants under the control of the native promoter, were lower than those of the same proteins expressed under the control of the 35S promoter. To obtain a greater amount of proteins and protein-containing complexes, we used HYL1D12-R1 and HYL1-R1 plants for the subsequent analysis of pre-miRNA processing.
Two N-Terminal dsRBDs Are Essential for miRNA Accumulation
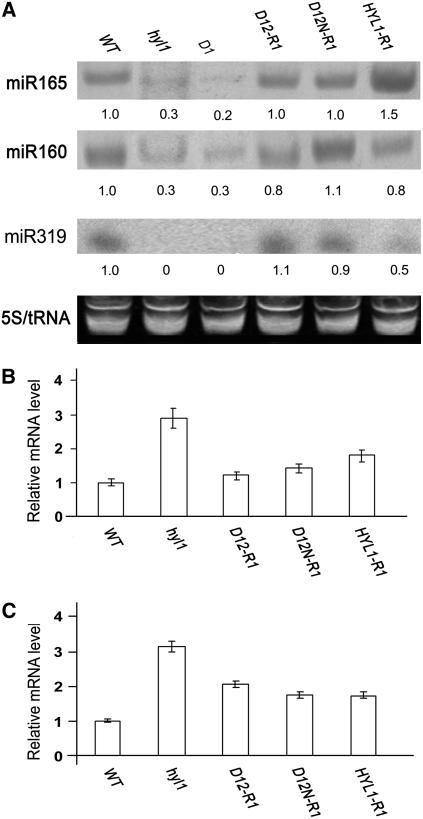
We detected the accumulation of miR165, miR160, and miR319 in D1, D12-R1, and D12N-R1 plants. Accumulation of the three miRNAs in the seedlings of D12-R1 and D12N-R1 plants was abundant and approximately the same as that seen in HYL1-R1 plants (Figure 4A). Compared with wild-type plants, accumulation of miR165 and miR160 in hyl1 mutant plants was much weaker, whereas that of miR319 was below detection. In the D1 plants, miR165 and miR160 accumulation was weak and miR319 accumulation was negligible, similar to that of hyl1 seedlings; notably, D12-R1 and D12N-R1 plants consistently accumulated the same amount of miR165, miR160, and miR319 as wild-type plants. These results suggest that the N-terminal dsRBDs in D12-R1 and D12N-R1 plants are capable of causing miR165 and miR160 accumulation. By contrast, the first N-terminal dsRBD alone is not sufficient to yield miRNA, as seen in D1 plants.
Figure 4.
Accumulation of miRNA and Expression of the Targeted Genes.
(A) Accumulation of miR165, miR160, and miR319 in seedlings of the transgenic lines D12-R1 and D12N-R1. Wild-type, hyl1, and HYL1-R1 plants were used as controls. Shown is an RNA gel blot analysis of 30 μg of total RNA prepared from a 2-week-old seedling with a DNA probe complementary to miR165. 5S rRNA and tRNA were used as loading controls. Normalized values of miR165, miR160, and miR319 to 5S rRNA/tRNA are indicated, with miRNA levels in ecotype Nossen set at 1.0.
(B) and (C) Expression of REV (B) and ARF17 (C) in mature rosette leaves. Real-time quantitative PCR quantifications normalized to the expression of UBQ5 are shown. D1, D12-R1, D12N-R1, and HYL1-R1 are the transgenic lines expressing Pro35S:HYL1D1, Pro35S:HYL1D12, Pro35S:HYL1D12N, and Pro35S:HYL1, respectively. hyl1 mutants and wild-type plants were used as the negative and positive controls, respectively. Error bars represent sd of three biological replicates.
The N-Terminal dsRBDs Cause the Reduced mRNA Levels of the Targeted Genes
One of the genes targeted by miR165/166 is REV, which regulates leaf polarity, and a target of miR160 is ARF17, which controls the auxin response. Our previous study indicated that the mutation of HYL1 causes increased REV expression (Yu et al., 2005). To determine whether or how the N-terminal dsRBDs of HYL1 are sufficient to cleave targeted mRNAs through a miRNA-directed pathway, we performed real-time quantitative PCR to detect the expression levels of REV and ARF17.
REV was expressed weakly in the leaves of D12-R1 and D12N-R1 plants, comparable to the level of expression seen in hyl1 mutants (Figure 4B). As the primers for REV were designed to encompass the fragment fully complementary to miR165, the PCR products should include the cleavage site within REV and thus should correspond only to the uncleaved transcripts. The weak expression of REV in D12-R1 plants indicates that N-terminal dsRBDs restore the function of HYL1, allowing the normal cleavage of REV mRNA.
For the analysis of ARF17 transcripts that are uncleaved by miR160, a pair of primers for real-time quantitative PCR was designed to surround the cleavage site; thus, the uncleaved transcripts were distinguished from the cleaved transcripts. ARF17 transcripts were less abundant in D12-R1 and D12N-R1 leaves than in hyl1 leaves but more abundant than in wild-type leaves (Figure 4C), indicating that Pro35S:HYL1D12 and Pro35S:HYL1D12N led to reductions in the levels of uncleaved ARF17 mRNA. On the other hand, the rescue of Pro35S:HYL1D12 and Pro35S:HYL1D12N in the levels of ARF17 transcripts was not complete, as ARF17 transcripts were more abundant in D12-R1 and D12N-R1 leaves than in wild-type leaves. Although this different abundance in ARF17 level did not affect the complete rescue of the phenotype conferred by hyl1 in D12-R1 plants, it may cause a physiological change in the auxin response of D12-R1 plants to some extent.
Two N-Terminal dsRBDs Are Sufficient for miRNA Processing
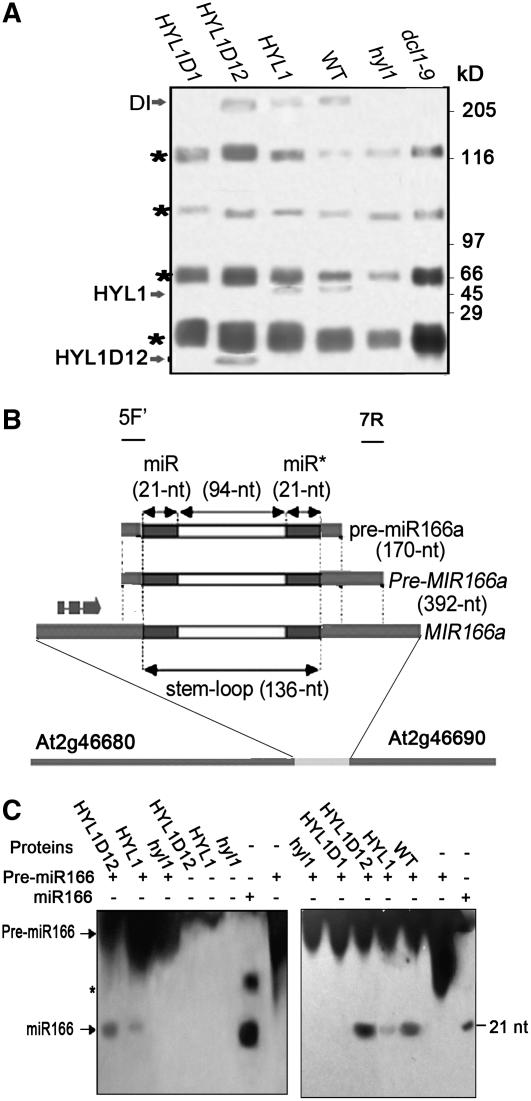
HYL1 interacts strongly with DCL1. However, it remains unknown whether HYL1-containing complexes from plant tissue are able to process pre-miRNA. Moreover, it is uncertain whether the N-terminal dsRBDs of HYL1 in transgenic plants can form a protein complex that is able to process pre-miRNA. To understand the function of the N-terminal dsRBDs, we studied the HYL1-containing complexes of transgenic plants using hyl1 mutants as a control in which the expression of the HYL1 gene was knocked down completely by Ds insertion. We raised polyclonal antibodies against HYL1D12N fragments of HYL1. The HYL1 antibody, which effectively immunoprecipitates HYL1D12 and HYL1 from extracts of different transgenic plants (data not shown), was used to identify the proteins associated with HYL1 in plant tissues. Proteins that were reproducibly copurified with HYL1 antibodies were retained. The immunoprecipitates were resolved by SDS-PAGE. As indicated in Figure 5A, immunoprecipitates derived from D12 plants contained HYL1D12 and a protein corresponding to DCL1 in size, still to be identified (DI), whereas immunoprecipitates from HYL1-R1 or wild-type plants contained HYL1 and DI proteins. By contrast, DI proteins were not detectable in hyl1 and dcl1-9 mutants.
Figure 5.
Pre-miRNA Processing.
(A) Isolation of HYL1-containing complexes. Plant proteins were isolated from extracts of the leaves of D1, D12-R1, HYL1-R1, and hyl1 plants. Immunoprecipitation was performed using anti-HYL1 antibody. Immunoprecipitates were fractionated by SDS-PAGE and visualized by silver staining. Arrows point to the bands containing HYL1, HYL1D12, and one protein corresponding to DCL1 in size, still to be identified (DI). The molecular masses of marker proteins are indicated at right. Asterisks denote contaminating polypeptides that were also present in the control immunoprecipitates from hyl1 extracts.
(B) Structure of the predicted pre-miR166 sequence relative to the annotated genes. 5F′ (166a-5F′) and 7R (166a-7R) indicate the positions of the forward and reverse primers for cloning of the Pre-MIR166a fragment with the 5′ end at the exact start of pre-miR166a and the 3′ end in the region after pre-miR166a.
(C) RNA gel blot to detect the processing of pre-miR166 after the miRNA processing assay using the immunoprecipitates of different transgenic plants. The concentration of immunoprecipitates added to the reaction solution visualized at left was twice that added to the reaction solution visualized at right. Twenty-one-nucleotide DNA and 21-nucleotide RNA of miR166 were used as marker miRNAs in the left and right panels, respectively. Arrows point to bands containing pre-miR166 and mature miR166. The molecular mass of the marker miRNA (nucleotides [nt]) is indicated at right. The asterisk denotes the secondary structure of the DNA marker of miR166.
For analysis of miRNA processing, a 392-nucleotide fragment of Pre-MIR166 was isolated by PCR twice from the genomic DNA of Arabidopsis using two pairs of the specific primers 166a-5F/166a-7R and 166a-5F′/166a-7R (Juarez et al., 2004) (Figure 5B). RNA samples of pre-miR166a was synthesized from Pre-MIR166a by T7 polymerase in vitro transcription and then used as the substrate for the processing reaction. Processing of pre-miR166 was performed as described previously (Denli et al., 2004; Haase et al., 2005). The addition of the HYL1- or HYL1D12-containing complexes to the reaction yielded the final, 21-nucleotide mature miRNA from pre-miR166a substrate, demonstrating that the N-terminal region of HYL1 containing two dsRBDs is capable of processing pre-miRNAs (Figure 5C). We compared the pre-miRNA processing activity of extracts prepared from different plant lines. The HYL1D12-containing complex extracted from the D12-R1 plants processed pre-miR166 RNA more efficiently than the complex extracted from wild-type plants. Extracts from hyl1 mutants were deficient in pre-miRNA processing, as the addition of the HYL1D1-containing complexes to the reaction did not generate any mature miR166.
The N-Terminal Region with dsRBDs Is Not Restricted to the Nucleus
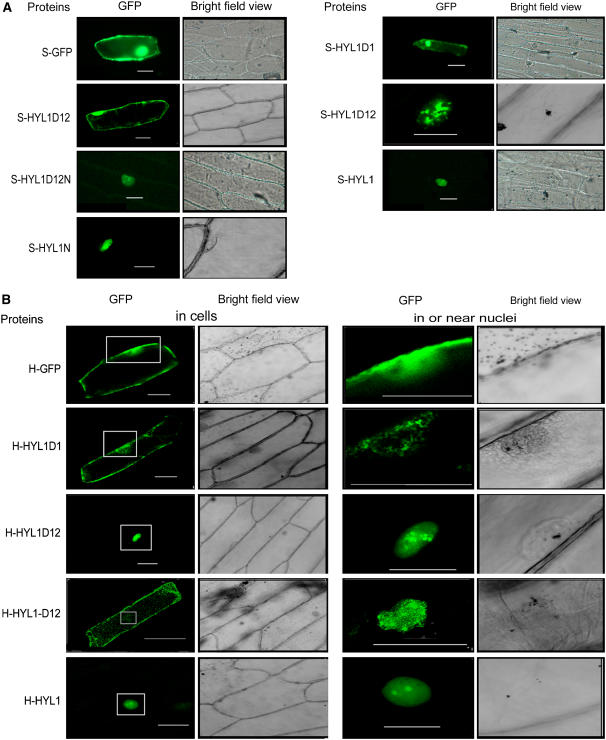
HYL1 was reported previously to be located in the nucleus, and the putative NLS domain is presumed to be responsible for nuclear localization (Lu and Fedoroff, 2000). To test whether the NLS domain of HYL1 is the true nuclear localization signal and where truncated HYL1 might be located in the absence of the NLS domain, we made constructs containing the various deletion mutants of HYL1 fused with GFP under the control of the 35S promoter: Pro35S:HYL1-GFP, Pro35S:HYL1D1-GFP, Pro35S:HYL1D12-GFP, and Pro35S:HYL1D12N-GFP. These constructs were introduced into onion (Allium cepa) epidermal cells with particle bombardment. In the cells bombarded with Pro35S:HYL1D12-GFP, the S-HYL1D12 protein was dispersed in the cytoplasm and nucleus in most epidermal cells with GFP fluorescence, similar to the pattern seen in cells expressing GFP only (Figure 6A). This protein was concentrated in the nucleus only in some cells (5 of 32 cells). By contrast, S-HYL1D12N was localized in the nucleus of all 47 cells examined for GFP fluorescence. Like S-HYL1, S-HYL1N, which corresponds to the NLS domain only, was localized in the nucleus, indicating that the NLS domain is indeed a nuclear localization signal.
Figure 6.
Subcellular Distribution of GFP-Fused Proteins.
(A) Subcellular localization of mutant protein variants of HYL1 produced under the control of the 35S promoter.
(B) Subcellular localization of mutant protein variants of HYL1 produced under the control of the native promoter.
Panels at right show a lower magnification of the areas surrounding the fluorescence signals in or near the nuclei (boxed) in the panels at left to show the precise distribution of the proteins. S-GFP, S-HYL1D1, S-HYL1-D12, S-HYL1D12N, S-HYL1, and S-HYL1N represent the proteins GFP, HYL1D1, HYL1-D12, HYL1D12N, HYL1, and HYL1N, respectively, generated under the control of the 35S promoter. H-GFP, H-HYL1D1, H-HYL1-D12, H-HYL1D12N, H-HYL1, and H-HYL1N represent the proteins GFP, HYL1D1, HYL1-D12, HYL1D12N, HYL1, and HYL1N, respectively, generated under the control of the native HYL1 promoter.
Considering that the subcellular localization of truncated HYL1 under the control of the 35S promoter might not represent the physiological situation, we then constructed HYL1 deletion mutants under the control of the native HYL1 promoter. In cells bombarded with ProHYL1:HYL1D12-GFP, H-HYL1D12 displayed a localization pattern similar to S-HYL1D12 in that it could be localized either in the nucleus or in both the nucleus and the cytoplasm (Figure 6B). These findings suggest that the N-terminal region containing two dsRBDs was not restricted to the nucleus. Unlike S-HYL1D1, H-HYL1D1 was seen only in the cytoplasm and was concentrated mainly around the nucleus. Apparently, S-HYL1D1 under the control of the 35S promoter was not restricted to the nucleus, but H-HYL1D1 under the control of the native promoter was restricted to the cytoplasm. The localization patterns of S-HYL1D12 and H-HYL1D12 were confirmed by localization analysis of truncated HYL1 in plants transgenic for the HYL1 deletion mutants (data not shown). We conclude that the putative NLS domain is the nuclear localization signal and that the N-terminal region containing the dsRBDs is not restricted to the nucleus.
DISCUSSION
HYL1 functions in the biogenesis and accumulation of miRNA, in combination with DCL1 and HEN1 (Han et al., 2004; Vaucheret et al., 2004; Vazquez et al., 2004a, 2004b). Like Pro35S:HYL1, both the Pro35S:HYL1D12 and ProHYL:HYL1D12 constructs of the HYL1D12 fragment completely rescue the phenotype conferred by hyl1. Corresponding to this phenotypic rescue, HYL1D12 (the N-terminal region containing two dsRBDs) in transgenic plants is able to form a complex with other specific proteins and to process pre-miRNA. One of the specific proteins copurified with HYL1 or HYL1D12 may be DCL1. DCL1 is considered to interact with the dsRBD2 of HYL1 (Hiraguri et al., 2005), an interaction that is required for the precise processing of pri-miRNA (Kurihara et al., 2006). On the basis of this interaction, we isolated the HYL1-containing complex from transgenic plants and addressed its role in the generation of mature miRNA. We show that the HYL1-containing complex purified from plant tissue is capable of processing pre-miRNA. Using the same methods, we demonstrate that the N-terminal region of HYL1 containing two dsRBDs of HYL1 is also sufficient to process pre-miRNA. Although the dsRBD2 was known previously to interact with DCL1, the elucidation of the physiological function of the two dsRBDs in miRNA processing advances our understanding of the mechanism of miRNA-directed silencing of many important genes. Specifically, this result suggests an important role for the N-terminal region of HYL1 and its two dsRBDs in facilitating miRNA processing. In contrast with the N-terminal region containing two dsRBDs, the first dsRBD of HYL1 failed to process pre-miRNA, indicating that both N-terminal dsRBDs are necessary for the processing of pre-miRNA.
HYL1 has been indicated to be present in the nucleus (Lu and Fedoroff, 2000; Han et al., 2004). However, the question remained whether the nuclear localization of HYL1 is determined by its putative NLS. NLS sequences, certain patterns of basic amino acid residues necessary for proteins to be imported into the nucleus, are classified as monopartite or bipartite. Proteins containing these signals are transported into nuclei by ATP-dependent translocation through the nuclear pore complexes (Richardson et al., 1988; Newmeyer and Forbes, 1990). The NLS sequence of HYL1 has a bipartite nuclear localization domain. In onion epidermal cells, the GFP fluorescence of HYL1-GFP and HYL1D12N-GFP containing the NLS domain was localized in the nucleus. By contrast, the fluorescence of HYL1D12-GFP without the NLS domain was seen in both nucleus and cytoplasm, whereas that of HYL1D1-GFP without the NLS and dsRBD2 was seen only in cytoplasm. This observation provides direct evidence that the putative NLS domain of HYL1 functions in nuclear localization. The random nuclear localization of HYL1D12, which lacks the NLS signal, may be related to SERRATE (SE), a general regulator of miRNA levels (Grigg et al., 2005; Lobbes et al., 2006; Yang et al., 2006). SE is localized in the nucleus, like HYL1, and interacts with HYL1. Yeast two-hybrid experiments show that both dsRNA binding motifs of HYL1 interact with SE (Lobbes et al., 2006). This interaction may account for the nuclear localization of HYL1 without an NLS. It is possible that SE randomly transports HYL1D12 into the nucleus using its own NLS signal, giving rise to random nuclear localization of HYL1D12.
Although the N-terminal region of HYL1 containing two dsRBDs lacks the NLS and PPI domains, it shows normal ability for miRNA processing, meaning that the NLS and/or the PPI domains of HYL1 are not essential for miRNA biogenesis. We consider that this N-terminal region acts either in the nucleus or in the cytoplasm. Tang et al. (2003) reported that standard wheat germ extract contains Dicer-like enzymes that convert dsRNA into two classes of small interfering RNA (miRNA and siRNA) and an endogenous miRNA complex that can direct the efficient cleavage of the wild-type Arabidopsis PHV mRNA sequence. Wheat germ extracts are essentially cytoplasm, and miRNA production in these extracts suggests the inclusion of HYL1 and HEN1 as partners of DCL1. Thus, we cannot exclude the possibility that miRNA biosynthesis as well as miRNA-guided cleavage of target mRNA may occur in the cytoplasm, where DCL1, HEN1, and HYL1 coexist. Alternatively, this protein variant may act only in the nucleus. Therefore, the question remains how this short protein is imported from the cytoplasm into the nucleus in the absence of an NLS domain, a process that may be related to other nuclear proteins, such as DCL1. dsRBD2 in the N-terminal region of HYL1 is able to form a complex with DCL1. In this way, DCL1 could bind to the N-terminal region of HYL1 containing two dsRBDs of HYL1 and bring it into the nucleus through the nuclear pore complex. The localization of the N-terminal region of HYL1 containing dsRBD1 but not dsRBD2 provides some evidence to support this possibility. As dsRBD1 lacks the ability to interact with DCL1, this fragment could not be brought into the nucleus. Under the control of the native promoter, the N-terminal region of HYL1 containing dsRBD1 stays in the cytoplasm, revealing a difference in subcellular localization from the N-terminal region of HYL1 containing both dsRBDs.
Plant development does not require full-length HYL1. D12-R1 plants of hyl1 mutants have the same phenotype as wild-type plants, indicating that the N-terminal dsRBDs play the same role in plant development as HYL1. In these plants, the mRNA levels of REV and ARF17 are reduced, whereas the accumulation of miR165 and miR160 is increased. This fact suggests that the N-terminal dsRBDs are involved in posttranscriptional gene silencing. In Drosophila, Dicer-1 requires Loquacious (Loqs) for efficient miRNA-mediated gene silencing (Forstemann et al., 2005). Loqs is a dsRNA binding protein with two dsRBDs homologous with dsRBD1 and dsRBD2 of HYL1. Along with Dicer-1, Loqs resides in a functional pre-miRNA processing complex and stimulates and directs the specific pre-miRNA processing activity, for which Loqs may directly bind Dicer-1 through its dsRBDs (Saito et al., 2005). It is interesting to speculate that HYL1 and Loqs have similar functions. In pre-miRNA processing, the N-terminal dsRBDs of HYL1 may bind DCL1, followed by the interaction between the featured domains of DCL1 and the N-terminal dsRBDs of HYL1. Recent studies have disclosed a complex containing TRBP, the homolog of Loqs in humans, and the association of the TRBP-Dicer complex with Argonaute2, the catalytic engine of RISC (Chendrimada et al., 2005; Haase et al., 2005). From these results, we propose that HYL1 or the N-terminal dsRBDs may play a similar role, not only in miRNA processing but also in RISC activity. Further insight into the mechanism by which HYL1 controls leaf polarity and the auxin response through miRNA mediation will largely depend on our understanding of the biochemical and physiological activities of the N-terminal dsRBDs.
METHODS
Plant Material and Growth Conditions
The hyl1 (Nossen ecotype) and dcl1-9 (Columbia ecotype) mutants of Arabidopsis thaliana were obtained from N.V. Fedoroff and J.C. Fletcher, respectively (Lu and Fedoroff, 2000; Williams et al., 2005). Seeds were surface-sterilized in 70% ethanol for 1 min, and then in 0.1% HgCl2 for 10 min, and washed four times in sterile distilled water. For tissue culture in vitro, the seeds were mixed in molten 0.1% water agar and plated on top of solid 1% sugar Murashige and Skoog medium. Plates were sealed with Parafilm, incubated at 4°C in darkness for 3 to 4 d, and then moved to a growth chamber at 22°C with 16 h of light. For phenotypic observation, seeds were sown in pots with peat soil and grown in growth chambers with the same conditions. For quantitative measurement of leaf incurvature (upward curvature) (u), the transverse curvature index (TC) is defined as TCu = (lm − pw)/pw, where lm is the distance between the lateral margins of incurved leaves and pw denotes pressed leaf width. For leaf downward curvature (d), the transverse curvature index is defined as TCd = (pw − lm)/pw. The fourth leaves on 25-d-old plants were cut transversely at the widest extreme and pressed on a desk for the measurement of pressed width when the distance between the lateral margins of the leaves at the widest extreme was measured.
Construction of HYL1 Deletion Mutants
The cDNA sequence of the HYL1 gene (AF276440) was originally isolated from a mixed cDNA library of Arabidopsis by PCR and cloned in pTA1 plasmids. The various deletion mutants of HYL1 were made by PCR, using cDNA of HYL1 as template. The primers we used include the forward primer HYL1-5 and the reverse primers HYL1D1-3, HYL1D12-3, HYL1D12N-3, and HYL1-3 (see Supplemental Table 2 online). A BamHI site at the 5′ end and a SalI site at the 3′ end were introduced for the construction of plant expression vectors. In the binary vector pJR1, the deletion fragments under the control of the cauliflower mosaic virus 35S promoter were constructed and verified for accuracy by DNA sequencing. Afterward, the HYL1 gene promoter sequence (850 bp) was amplified using the primer pHYL1-A1 with an EcoRI site at the 5′ end and pHYL1-S1 with a BamHI site at the 3′ site. For comparison, the deletion mutants were constructed under the control of the native promoter by replacing the 35S promoter with the native HYL1 promoter. All constructs, with either the 35S or the native promoter, were transferred into the binary vector pCAMBIA3301 of Agrobacterium tumefaciens. Genetic transformation of homozygous hyl1 mutant plants was performed using the floral dip method (Clough and Bent, 1998).
For selection of transgenic plants, seeds were sterilized and germinated on agar medium containing 50 mg/L kanamycin or 10 mg/L phosphinothricin. Seedlings resistant to both kanamycin and phosphinothricin were transplanted in a growth room and grown at 23°C under 12 h of light and12 h of dark. The transgenic plants D1, D12-R1, D12N-R1, and HYL1-R1 were identified using PCR and DNA gel blot hybridization and then self-fertilized for at least three generations. Seeds from each plant were harvested separately for subsequent observation.
Gene Expression Analysis
Total RNA was isolated from seedlings (4 weeks) and mature leaves of the transgenic plants (7 weeks) expressing the deletion mutants of HYL1. First-strand cDNA was synthesized for real-time quantitative PCR. Primers specific for the cDNA of UBIQUITIN5 (UBQ5) were used to normalize the amplification of sample DNA fragments (see Supplemental Table 2 online). The primers E-HYL1-5/E-HYL1D1-3 and E-HYL1-5/E-HYL1D12-3, specific for HYL1D1 and HYL1D12, respectively, were used to detect expression levels of the deletion mutants. The primers ARF17-5/ARF17-3 and REV-5/REV-3 were designed against each side of the cleavage site and used to detect uncleaved REV (AF233592) and ARF17 (At1g77850) transcripts, respectively. Real-time PCR was performed in the Rotor-Gene 3000 real-time PCR cycler using the SYBR Premix Ex Taq kit (Takara), as described previously (Yu et al., 2005). For each cDNA synthesis, quantification was performed in triplicate. For each quantification, a melt curve was realized at the end of the amplification experiment by steps of 0.5°C from 65°C to 95°C to ensure that quantification was not caused by primer self-amplification. Results were normalized to those for UBQ5, then to the expression level of the wild-type plants.
miRNA Isolation and Blot Analysis
Total RNA was extracted from seedlings of 2-week-old wild-type plants, hyl1 mutants, and transgenic plants. Fifteen micrograms of total RNA was fractionated on a 15% polyacrylamide gel containing 8 M urea and transferred to a Nitran Plus membrane (Schleicher and Schuell). Antisense sequences (21 bp) of miR165, miR160, and miR319 were synthesized and end-labeled as probes with [γ-32P]ATP using T4 polynucleotide kinase (Takara). Hybridization was performed at 41°C using hybridization buffer (7% SDS, 1% BSA, 1 mM EDTA, and 0.25 M NaHPO4, pH 7.2). A synthesized miR165 was used as a positive control, and tRNA and 5S rRNA were used for the quantity control of total RNA content between samples.
Protein Purification and Antibody Preparation
The HYL1D12N fragment of the HYL1 cDNA (without the PPI domain) was constructed in the expression vector pMAL-C2, and the recombinant plasmids were transformed into Escherichia coli DH5α. Bacteria were cultured at 37°C overnight in a 20-mL volume and then diluted to a 200-mL volume with 4 mM isopropylthio-β-galactoside and cultured at 37°C for 4 h. Nearly 2 liters of cultured bacteria was collected and lysed by ultrasonic precipitates, and the precipitates were then resolved in a buffer containing 1% Triton X-100, 20 mM Tris, 20 mM EDTA, and 1 mM DTT. The supernatant was purified using an amylase-prepacked column (New England Biolabs) and washed with a buffer containing 5 M NaCl. Rabbit antibody against HYL1D12N was obtained by immunizing a rabbit with the purified HYL1D12N protein. The titer and specificity of the antibody were determined by ELISA and protein gel blot analysis, respectively.
Protein Gel Blot Analysis
Protein samples were analyzed on 12% SDS–polyacrylamide gels and transferred to an Immobilon-P membrane (Millipore Intertech). Membranes were blocked with 5% nonfat dry milk in PBS plus 0.2% Tween 20 (PBST), then probed with rabbit polyclonal anti-HYL1 antibodies in PBST that were raised against maltose binding protein recombinant HYL1 protein expressed in E. coli. Primary antibodies were detected using a horseradish peroxidase–labeled goat anti-rabbit IgG secondary antibody (Chemicon). Bands were visualized by an enhanced chemiluminescence system according to the manufacturer's instructions (Chemicon).
Transient Expression Assay of Protein Variants of HYL1 in Plant Cells
The 35S-HYL1 deletion fragments for GFP fusion were produced by amplifying the HYL1 coding sequence with the forward primer G-HYL1-5 and the following reverse primers: G-HYL1D1-3, G-HYL1D12-3, G-HYL1D12N-3, and G-HYL1-33 (see Supplemental Table 2 online). Amplified fragments were digested with BamHI and KpnI and cloned in frame with the N terminus of the GFP open reading frame in the plasmid pEGFP digested with BamHI/KpnI. Deletion fragments fused in-frame to GFP were subcloned into the pJR1 binary vector, and the fused HYL1-GFP constructs were placed under the control of the cauliflower mosaic virus 35S promoter by BamHI/XbaI. All fusion constructs were verified for accuracy by DNA sequencing. Onion (Allium cepa) epidermal layers were transformed using biolistic bombardment as described by Varagona et al. (1992). After bombardment, the layers were incubated for 20 h at room temperature in darkness. The subcellular location of the green fluorescent precipitate was transiently assayed with a confocal microscope. Meanwhile, HYL1 deletion mutants fused with GFP under the control of the 35S promoter or the native promoter were transformed into hyl1 mutants and wild-type plants of Arabidopsis, and the subcellular location of GFP in the transgenic plants was observed.
Isolation of the HYL1-Containing Complex
Plants were ground into a fine powder using liquid nitrogen and then resolved in a buffer containing 20 mM Tris-HCl, pH 7.4, 200 mM KCl, 40 mM MgCl2, 1 mM DTT, 20% glycerol, and 0.05% Nonidet P-40. To purify the HYL1-containing complexes, cell extracts generated from the different transgenic lines of Arabidopsis were incubated with 20 μL of Protein A–Sepharose (Amersham Biosciences) at 4°C for 1 h. Precleared extracts were then incubated with 5 μL of anti-HYL1 serum for 1 h and then 20 μL of Protein A–Sepharose at 4°C for 1 h. Immunoprecipitates were washed three times in extraction buffer (20 mM Tris-HCl, pH 7.9, 0.5 M KCl, 10% glycerol, 1 mM EDTA, 5 mM DTT, 0.5% Nonidet P-40, and 0.2 mM phenylmethylsulfonyl fluoride).
Preparation of RNA Substrates and RNA Processing Assays
The Pre-MIR166a fragment of 392 nucleotides was obtained by amplification of the genomic DNA of Arabidopsis using two pairs of primers, 166a-5F/166a-7R and 166a-5F′/166a-7R, and inserted in pGEM-7Z vector. The pre-miR166a fragment, which starts at the exact 5′ end of the predicted pre-miR166a and extends to 222 bp downstream of pre-miR166a, was generated by in vitro transcription using linearized pGEM-7Z vector. It was synthesized by T7 polymerase in vitro transcription using an Ambion T7 MaxiScript transcription kit. After transcription, samples were treated with DNase I, extracted with phenol, and RNA-purified by denaturing 8% PAGE. After dephosphorylation by calf intestine phosphatase, RNAs were 5′ end–phosphorylated using T4 polynucleotide kinase and ATP. Complementary RNA strands were annealed at 95°C for 3 min in 20 mM NaCl, transferred to 75°C, and then slowly cooled to 20°C. Pre-miR166 RNA was dissolved in water and renatured by incubation at 90°C for 1 min, followed by incubation at 25°C for 15 min in 30 mM Tris-HCl, pH 6.8, containing 50 mM NaCl, 2 mM MgCl2, and 10% glycerol.
The processing reaction consisted of the indicated amounts of the HYL1 complex, 3 mL of solution containing 32 mM MgCl2, 10 mM ATP, 200 mM creatine phosphate, 30 μg/mL creatine kinase, and 1 unit/μL human placental ribonuclease inhibitor (Takara), and the pre-miRNA. Processing buffer (20 mM Tris-HCl, pH 7.9, 0.1 M KCl, 10% glycerol, 5 mM DTT, and 0.2 mM phenylmethylsulfonyl fluoride) was added to a final volume of 30 mL. The reaction mixture was incubated at 37°C for 90 min and extracted with a phenol:chloroform mixture, and then with chloroform, and precipitated with 300 mM sodium acetate and ethanol. Precipitated RNA was loaded and resolved on 19% denaturing PAGE gels and transferred to Hybond N+ membranes (Amersham). Oligonucleotide probes (5′-TCGGACCAGGCTTCATTCCCC-3′) complementary to miR166 were 5′ end–labeled using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. Prehybridization and hybridization solutions contained 2× SSPE (1× SSPE is 0.115 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA, pH 7.4), 5× Denhardt's solution (1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), 0.1% SDS, and 20% formamide. After prehybridization for 3 h, hybridization with the labeled probes was performed at 37°C for at least 5 h. Membranes were washed five times with 2× SSPE and 0.2% SDS. To determine miRNA abundance in different transgenic lines, two independent RNA preparations were analyzed in quadruplicate.
Accession Numbers
Accession numbers (GenBank/EMBL or Arabidopsis Genome Initiative) of the major genes discussed in this article are as follows: HYL1 (At1g09700), REV (At5g60690), and ARF17 (At1g77850).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Diagrams of the Constructs with Deletion Mutants of the HYL1 Gene under the Control of the Native Promoter of HYL1.
Supplemental Table 1. Phenotype Rescue of hyl1 Mutants by Deletion Mutants of HYL1 within T1 Populations.
Supplemental Table 2. Primer Sequences.
Supplementary Material
Acknowledgments
We thank N.V. Fedoroff and J.C. Fletcher for providing the seeds of hyl1 and dcl1-9 mutants, respectively. This work was supported by grants from the Natural Science Foundation of China (Grants 39870450 and 90508005) and the High Tech Research and Development Project of China (Grant 2006AA02C139).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yuke He (ykhe@sibs.ac.cn).
Online version contains Web-only data.
References
- Ambros, V. (2004). The functions of animal microRNAs. Nature 431 350–355. [DOI] [PubMed] [Google Scholar]
- Bandziulius, R.J., Swanson, M.S., and Dreyfuss, G. (1989). RNA-banding proteins as developmental regulators. Genes Dev. 3 431–437. [DOI] [PubMed] [Google Scholar]
- Bartel, B., and Bartel, D.P. (2003). MicroRNAs: At the root of plant development? Plant Physiol. 132 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409 363–366. [DOI] [PubMed] [Google Scholar]
- Boutet, S., Vazquez, F., Liu, J., Beclin, C., Fagard, M., Gratias, A., Morel, J.B., Crete, P., Chen, X., and Vaucheret, H. (2003). Arabidopsis HEN1: A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada, T.P., Gregory, R.I., Kumaraswamy, E., Norman, J., Cooch, N., Nishikura, K., and Shiekhattar, R. (2005). TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436 740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Denli, A.M., Tops, B.B., Plasterk, R.H., Ketting, R.F., and Hannon, G.J. (2004). Processing of primary microRNAs by the Microprocessor complex. Nature 432 231–235. [DOI] [PubMed] [Google Scholar]
- Forstemann, K., Tomari, Y., Du, T., Vagin, V.V., Denli, A.M., Bratu, D.P., Klattenhoff, C., Theurkauf, W.E., and Zamore, P.D. (2005). Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 3 e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg, S.P., Canales, C., Hay, A., and Tsiantis, M. (2005). SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature 437 1022–1026. [DOI] [PubMed] [Google Scholar]
- Haase, A.D., Jaskiewicz, L., Zhang, H., Laine, S., Sack, R., Gatignol, A., and Filipowicz, W. (2005). TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 6 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M.H., Goud, S., Song, L., and Fedoroff, N.V. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. USA 101 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraguri, A., Itoh, R., Kondo, N., Nomura, Y., Aizawa, D., Murai, Y., Koiwa, H., Seki, M., Shinozaki, K., and Fukuhara, T. (2005). Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 57 173–188. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., Running, M.P., and Meyerowitz, E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126 5231–5243. [DOI] [PubMed] [Google Scholar]
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, B.A., and Timmermans, M.C. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 84–88. [DOI] [PubMed] [Google Scholar]
- Kurihara, Y., Takashi, Y., and Watanabe, Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12 206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., and Kim, V.N. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425 415–419. [DOI] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lobbes, D., Rallapalli, G., Schmidt, D.D., Martin, C., and Clarke, J. (2006). SERRATE: A new player on the plant microRNA scene. EMBO Rep. 7 1052–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., and Fedoroff, N.V. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12 2351–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, E., Guttinger, S., Calado, A., Dahlberg, J.E., and Kutay, U. (2004). Nuclear export of microRNA precursors. Science 303 95–98. [DOI] [PubMed] [Google Scholar]
- Newmeyer, D.D., and Forbes, D.J. (1990). An N-ethylmaleimide-sensitive cytosolic factor necessary for nuclear protein import: Requirement in signal-mediated binding to the nuclear pore. J. Cell Biol. 110 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp, I., Mette, M.F., Aufsatz, W., Daxinger, L., Schauer, S.E., Ray, A., Van der Winden, J., Matzke, M., and Matzke, A.J. (2003). Evidence for nuclear processing of plant microRNA and short interfering RNA precursors. Plant Physiol. 132 1382–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, W.D., Mills, A.D., Dilworth, S.M., Laskey, R.A., and Dingwall, C. (1988). Nuclear protein migration involves two steps: Rapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell 52 655–664. [DOI] [PubMed] [Google Scholar]
- Ryter, J.M., and Schultz, S.C. (1998). Molecular basis of double-stranded RNA-protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., Ishizuka, A., Siomi, H., and Siomi, M.C. (2005). Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 3 e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer, S.E., Jacobsen, S.E., Meinke, D.W., and Ray, A. (2002). DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 7 487–491. [DOI] [PubMed] [Google Scholar]
- Schwab, R., Palatnik, J.F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8 517–527. [DOI] [PubMed] [Google Scholar]
- St. Johnston, D., Brown, N.H., Gall, J.G., and Jantsch, M. (1992). A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. USA 89 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona, M.J., Schmidt, R.J., and Raikhel, N.V. (1992). Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez, F., Gasciolli, V., Crete, P., and Vaucheret, H. (2004. a). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14 346–351. [DOI] [PubMed] [Google Scholar]
- Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A.C., Hilbert, J.L., Bartel, D.P., and Crete, P. (2004. b). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16 69–79. [DOI] [PubMed] [Google Scholar]
- Williams, L., Grigg, S.P., Xie, M., Christensen, S., and Fletcher, J.C. (2005). Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132 3657–3668. [DOI] [PubMed] [Google Scholar]
- Yang, L., Liu, Z., Lu, F., Dong, A., and Huang, H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47 841–850. [DOI] [PubMed] [Google Scholar]
- Yi, R., Qin, Y., Macara, I.G., and Cullen, B.R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L., Yu, X.H., Shen, R.J., and He, Y.K. (2005). HYL1 gene maintains venation and polarity of leaves. Planta 221 231–242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.