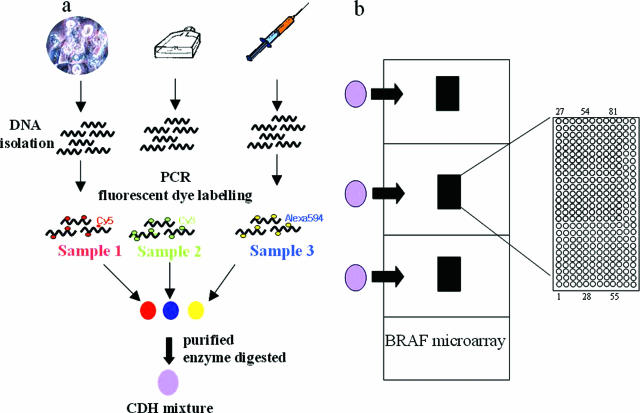
Figure 1.
Schematic depiction of CDH analysis of our BRAF oligonucleotide microarray with three samples. a: DNA was extracted from a cancer tissue sample, a cell line, and blood. The isolated DNA was amplified with three different fluorescent dyes (Cy5, Cy3, and Alexa-594), and the fluorescently labeled PCR products were mixed, purified, enzyme digested, and concentrated. The pellet was then resuspended in hybridization solution and hybridized to the spotted areas. b: Our BRAF oligonucleotide microarray contained three copies of the full array on one slide, allowing three hybridizations per slide. Because CDH allowed hybridization of three different samples per area, this technique could allow hybridization of nine total samples per microarray. The enlarged spotted area shows that a total of 324 (81 oligonucleotides in quadruplicate) were spotted per area. Each oligonucleotide was printed four times horizontally, in an upward direction starting from position 1, and the printing was divided into three groups. Positions 1 to 27 were printed first, then positions 28 to 54, and finally, positions 55 to 81.