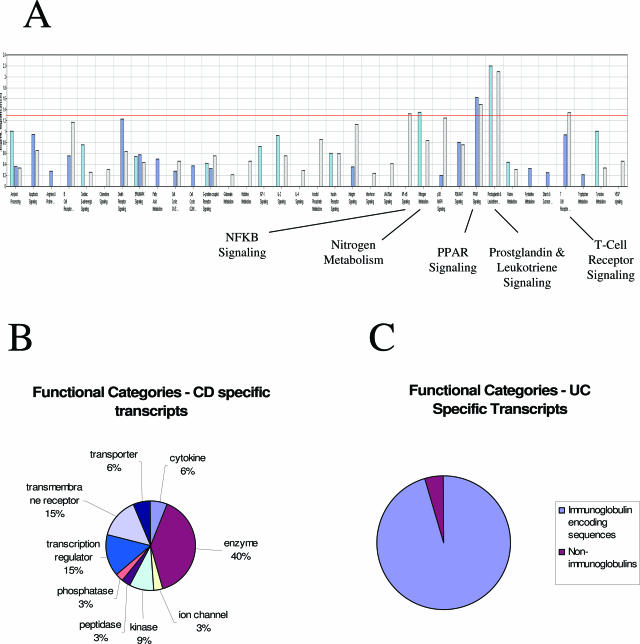
Figure 1.
Functional annotation and categories of transcripts identified as CD-associated, UC-associated, and differentially expressed between UC and CD. A: Canonical pathways overrepresented in the CD versus normal analysis of covariance comparison (light blue); the UC versus normal analysis of covariance comparison (dark blue), and the UC versus CD analysis of covariance comparison (light purple). In this panel the negative log10 of the P value is plotted to highlight more significant associations. The pathways interrogated (in order) were as follows: amyloid processing, apoptosis signaling, arginine and proline metabolism, B-cell receptor signaling (nonimmunoglobulin), cardiac β-adrenergic signaling, chemokine signaling, death receptor signaling, ERK/MAPK signaling, fatty acid metabolism, cell cycle regulation (G1/S), cell cycle regulation (G2/M), GPCR signaling, glutamate metabolism, histidine metabolism, IGF-1 signaling, IL-2 signaling, IL-4 signaling, inositol phosphate metabolism, insulin receptor signaling, integrin signaling, interferon signaling, JAK/STAT signaling, NF-κB signaling, nitrogen metabolism, p38 MAPK signaling, PI3K/AKT signaling, PPAR signaling, prostaglandin and leukotriene metabolism, purine metabolism, pyrimidine metabolism, starch and sucrose metabolism, T-cell receptor signaling, tryptophan metabolism, tyrosine metabolism, and VEGF signaling. B: CD-specific transcripts in PBMCs were functionally annotated using the Ingenuity pathway analysis system (Ingenuity), and the relative distribution of transcripts in each of the chosen functional categories for the CD-associated genes are presented in the pie chart. C: UC-specific transcripts in PBMCs were functionally annotated manually, and the relative distribution of transcripts in immunoglobulin versus nonimmunoglobulin categories is presented.