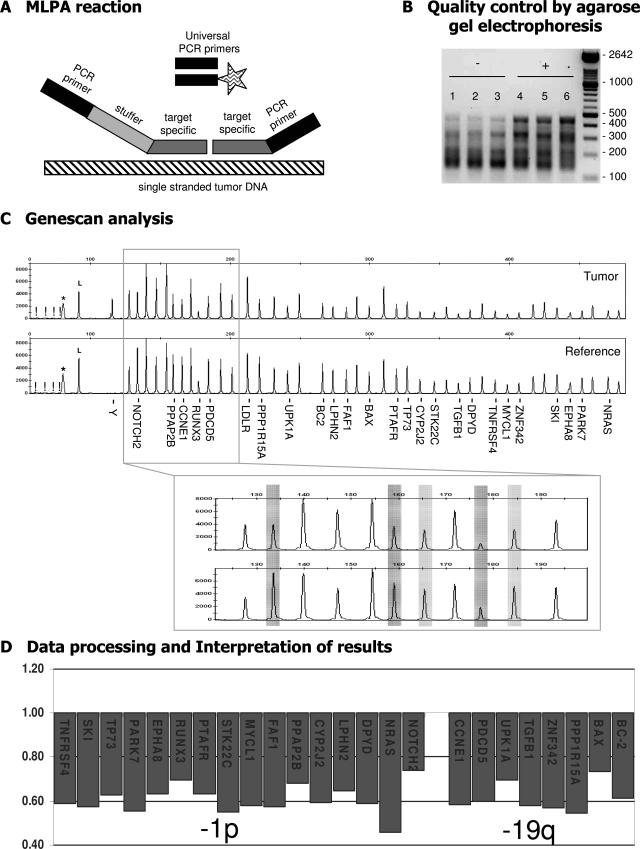
Figure 1.
Schematic outline of the MLPA procedure. During MLPA analysis up to 45 target-specific probes are used in one reaction. A: The unique target-specific sequences (gray) of the probes are hybridized on single-stranded genomic DNA (hatched) after which the two adjacent parts of the probe are joined through ligation and PCR amplification is performed with a fluorescently (FAM) labeled primer (indicated by the asterisk). Because identical PCR primers (black) are used for all probes, there are no primer-specific advantages in this procedure. Each probe has a unique length [attributable to variation in the length of the stuffer sequence (light gray)], and therefore gel electrophoresis can be used to separate the individual probe fragments. B: Agarose gel electrophoresis is performed as an early (and extra) control step for identification of suboptimal MLPA reactions (P088 is shown as an example). DNA fragments are smaller than 500 bp. In suboptimal samples (−; lanes 1 to 3) the large DNA fragments are underrepresented compared to adequate experiments (+; lanes 4 to 6). C: Capillary electrophoresis is performed to identify and quantify the amplification products. Probe names are provided for those located on 1p and 19q. The MLPA mix includes an internal control for DNA quantity and quality. If these are insufficient, fragments of 64, 70, 76, and 82 bp will appear (indicated by an exclamation mark in C) whereas a band of 94 bp indicates successful ligation and hybridization (indicated by an L in C). A nonspecific broad peak can be present, with the size depending on the electrophoresis apparatus and fluorescence used (indicated by an asterisk in C). In this example, MLPA analysis with kit P088 of an oligodendroglial tumor (N293) with −1p/−19q (as detected by CGH) is shown. Subsequently, ratio analysis is performed, and results are visualized showing the probes in their chromosomal order (described in Materials and Methods; threshold used for losses and gains set at 0.8 and 1.2, respectively).