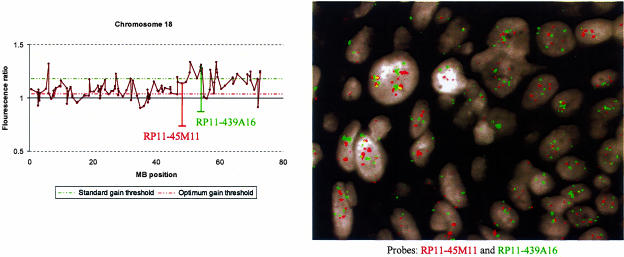
Figure 5.
Comparison of optimized thresholds with standard thresholds when applied to array CGH data generated from a frozen primary cervical SCC sample known to be hypertriploid or tetraploid. The gain threshold optimized to hypertriploid samples was 1.04 whereas the standard +3 SD gain threshold was 1.18. The ratio profile of chromosome 18, together with interphase FISH on cells of the same SCC sample, are illustrated. The optimum gain threshold detects gain of both clones RP11-45M11 and RP11-439A16, whereas the standard gain threshold fails to detect gain of RP11-45M11. FISH on interphase nuclei confirms gain of RP11-45M11 (red) as well as RP11-439A16 (green).