Abstract
The main goal of this study was to obtain new isolates of Leptospira spp. from sheep. A total of ten kidney samples and 44 blood samples were collected from sheep slaughtered in Pelotas, Southern Brazil. One isolate was obtained which was identified by 16S rRNA gene sequencing and serogrouping to be Leptospira noguchii serogroup Autumnalis. Microscopic agglutination test (MAT) evaluation revealed that 4.5% of the sheep sera reacted against the Autumnalis serogroup. This is the first report of isolation of L. noguchii from sheep. Together these findings indicate that L. noguchii infections may be a potentially important veterinary problem in this domestic animal species.
Keywords: leptospirosis, Leptospira noguchii, isolation, serogrouping, sheep, Brazil
Introduction
Leptospirosis in sheep has been reported in all continents over the years and infection with different leptospiral serovars have been reported from different countries (Hathaway et al., 1982; Faine et al., 1999; Ciceroni et al., 2000). Sheep may acquire the disease from contaminated urine of rodents, cattle, or other farm animals. Earlier isolation of L. interrogans serovar Hardjo (Bahaman et al., 1980; Ellis et al., 1983; Cousins et al., 1989; Gerritsen et al., 1994), and L. borgpetersenii serovar Javanica (Natarajaseenivasan and Ratnam, 1999), indicates that this host species may act as an important reservoir disseminating leptospires to both, animals and humans.
In Brazil, little has been reported concerning leptospiral isolation and its role in the etiology of leptospirosis in sheep. Azevedo et al. (2004) reported isolation of leptospires from sheep kidneys without clinical signs of leptospirosis in Northeast region of Brazil; however, the species were not identified. Serological studies suggest that the most prevalent serogroups associated with sheep worldwide are Sejroe, Grippotyphosa, and Pomona (Faine et al., 1999). In South America, Ciceroni et al. (1997) demonstrated a seropositive rate of 14.3% and serovars Poi and Pomona, were the most prevalent. In Chile, Zamora et al. (1999) detected a seropositivity of 5.7% and serovars Icterohaemorrhagiae, Autumnalis, and Hardjo appeared as the most prevalent. In Brazil, studies performed by Santa Rosa and Castro (1963), Favero et al. (2002), and Herrmann et al. (2004) demonstrated as the most prevalent serogroups Sejroe, Hebdomadis, Autumnalis, Pyrogenes, Australis, Ballum, Pomona, Icterohaemorrhagiae, and Grippotyphosa, but in these studies no leptospires were isolated. In this context, the purpose of the present study was to obtain and characterize isolates of Leptospira spp. from sheep kidney tissue collected from a slaughterhouse in Pelotas, Southern Brazil and to perform a serological survey to evaluate the prevalence among the slaughterhouse sheep population.
Methods
Animals and samples
Sheep used in this study comprised of apparently healthy animals slaughtered in a municipal abattoir called Frigorífico Caco located in the vicinity of Pelotas, Rio Grande do Sul State, Brazil. Kidneys from ten animals and serum samples from forty-four animals were obtained during four visits between May and October, 2003.
Isolation procedure
Each kidney was aseptically removed immediately after slaughter, and transported to the laboratory under refrigerated conditions in individual sterile polypropylene bags. In the laboratory, tissue samples were taken by perforation with a sterile Pasteur pipette after superficial cauterization, and inoculated in tubes containing 5 ml of Ellinghausen-McCullough-Johnson-Harris liquid medium (EMJH) (Difco-USA) with the addition of 10% rabbit serum, without antibiotics. Cultures were incubated at 29°C and examined weekly, during 10 weeks, by dark field microscopy. When growth was detected, successive transfers were made in liquid and semisolid media until growth was sufficiently abundant.
Virulence determination
To determine if the isolate would produce infection in laboratory animals, a group of four 28-day-old hamsters were inoculated intraperitoneally with 108 leptospires in a final volume of 1 ml. Animals were confined in isolator cages and monitored daily for the presence of clinical signs, including evidence of external hemorrhage, dehydration, ruffled hair coat, decreased activity and isolation. Four weeks after inoculation all hamsters were euthanized and the kidneys were aseptically removed, macerated and suspended in liquid media for reisolation (Faine, 1982). All animal procedures carried out in this study were approved by the Committee for Animal Care and Use (CEUA/FIOCRUZ, License 035).
Serogrouping and serologic testing
Microscopic agglutination test (MAT) was performed to serogroup the Leptospira isolate using rabbit antisera against reference serovars representing a standard battery of 21 pathogenic serogroups (Faine, 1982). Confirmation of serogroup status was performed by raising hyperimmune antisera against the Leptospira isolate in New Zealand White rabbits. Hyperimmune sera were evaluated by MAT against a battery of 36 reference strains that represented 26 serovars to determine whether maximum agglutination titers recognized serovars within the presumed serogroup. For the slaughterhouse survey of leptospiral infection, serum samples from sheep were evaluated in the MAT using a panel of 49 serovars (WHO, 2003). A positive MAT reaction was defined as an agglutination titer of 1:100 or greater.
DNA extraction and 16S rRNA gene sequencing
Genomic DNA was extracted using the GFX Genomic Blood DNA Purification Kit following the protocol for Gram-negative bacteria recommended by the manufacturer (GE healthcare). The extracted DNA was submitted to agarose gel electrophoresis in order to quantify and evaluate its integrity and quality, and stored at − 20°C. The whole 16S rRNA gene was amplified with the universal primers (Weisburg et al. 1991) and sequenced using internal primers designed in the present study, F2 - GGCGGCGCGTCTTAAACATG, F4 - GTGCCAGCAGCCGCGGTAA, F6 – AGTGAACGGGATTAGATACC and R11 – CCTAGACATAAAGGCCATGA. Fragments were amplified with one cycle at 94°C for 3 min, 35 cycles at 94°C for 30 sec, 52°C for 30 sec, 72°C for 1.5 min and a final extension at 72°C for 7 min. Aliquots were evaluated by agarose gel electrophoresis. Before the sequencing step, PCR products were purified by the use of GFX PCR DNA and Gel Band purification kit according to manufacturer instructions (GE Healthcare). The sequencing was performed in a MegaBACE 500 DNA sequencer (GE Healthcare) by the use of the Dynamic ET-terminator technology. Chromatograms were assembled and analysed using ContigExpress® module of Vector NTI 10.0 suite (Invitrogen). The assembled sequence was submitted to BLAST alignment (www.ncbi.nlm.nih.gov/BLAST) against other 16S rRNA sequences available in GenBank. The 16S rRNA sequence of Caco strain was deposited in GenBank under the accession number DQ848350.
Phylogenetic analysis
The 16S rRNA sequences available in GenBank from 11 strains (Table 1), comprising the members of the Autumnalis serogroup, plus members of the L. noguchii species were used in order to assemble a phylogenetic tree with the MEGA 3.1 software (Kumar et al., 2004). One thousand bootstrap replications were used for the correct positioning of the strains into the branches. The assembled tree was constructed by the neighbor-joining method and the results were given regarded to p-distance.
Table 1.
Strains used in the phylogentic analysis
| Species | Serogroup | Serovar | Strain | Accession number |
|---|---|---|---|---|
| L. interrogans | Autumnalis | Autumnalis | Akiyami A | AY996791 |
| L. interrogans | Autumnalis | Bulgarica | Mallika | AY996792 |
| L. kirschneri | Autumnalis | Erinaceiauriti | Erinaceus Auritus 670 | AY461874 |
| L. kirschneri | Autumnalis | Bim | 1051 | AY996802 |
| L. borgpetersenii | Autumnalis | Srebarna | 1409/69 | AY461855 |
| L. noguchii | Autumnalis | - | Caco | DQ848350 |
| L. noguchii | Autumnalis | Fortbragg | Fort Bragg | U12671 |
| L. noguchii | Panama | Panama | CZ214K | AY631886 |
| L. noguchii | Australis | Nicaragua | 1011 | AY461880 |
| L. noguchii | Louisiana | Orleans | LSU2580 | AY461881 |
| L. noguchii | Pomona | Proechimys | LT796 | AY461882 |
Results
Leptospira isolation was obtained from one out of 10 randomly selected sheep kidneys. The isolate named strain Caco showed slow growth, requiring 6 weeks of incubation before leptospires could be detected by dark field microscope examination. Strain Caco was recovered from kidneys of experimentally inoculated hamsters 4 weeks after inoculation. However, no clinical signs of leptospirosis were observed in any of the inoculated animals. Serogrouping of the isolate revealed agglutination titers of 1:25,600, 1:3,200 and 1:200 for antisera against serogroups Autumnalis, Grippotyphosa and Pomona, respectively, and negative reactions with antisera against the remaining 18 serogroups. Furthermore, hyperimmune sera against strain Caco agglutinated all strains tested which belonged to serogroup Autumnalis. Agglutination titers were 1:25,600 against the strain Caco isolate, and 1:6,400 and 1:800 against reference strains Akiyami A and Rachmat, respectively. There were no detectable agglutination reactions with the 34 reference strains belonging to non-Autumnalis serogroups.
For species identification, the 16S rRNA gene was amplified and sequenced. A 1,394 bp sequence was obtained and used for BLAST alignment. Strain Caco demonstrated 99% identity (1390/1394 bp) with L. noguchii strain Fort Bragg, which also belongs to serogroup Autumnalis. To demonstrate a more complete taxonomic classification, a phylogenetic tree was assembled (Figure 1). A 1,182 bp sequence from several Leptospira strains comprising members of the Autumnalis serogroup or L. noguchii species (Table 1) was used. In agreement with BLAST results, strain Caco was closely related to L. noguchii strains, especially with strain Fort Bragg (Autumnalis), 1011 (Australis) and LSU2580 (Louisiana). Strains CZ214K and LT796, which belong to Panama and Pomona serogroups, respectively, were more distantly related. Members of the Autumnalis serogroup but belonging to other Leptospira species were positioned in separate branches. These data confirms Caco as belonging to L. noguchii species.
Figure 1.
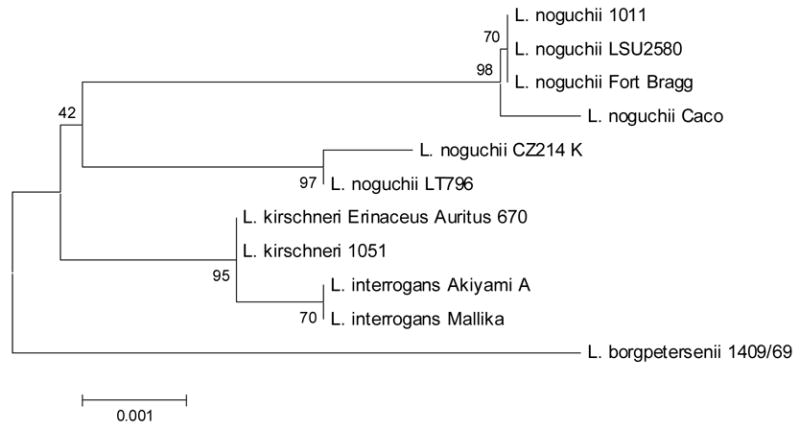
Phylogenetic position of Caco strain based on the 16S rRNA sequence. This phylogenetic tree summarizes, by bootstrap-based topology, the evolutionary relationship among L. noguchii strains and among members of other leptospira species belonging to the Autumnalis serogroup. The bootstrap consensus values are indicated over each root.
The survey found that 20.5% (9) of the 44 sheep in the slaughterhouse had MAT evidence of prior leptospiral infection. Serum samples had agglutination reactions against reference strains belonging to five different serogroups (Table 2). Two out of nine positive animals had agglutinating antibodies against serogroup Autumnalis strains. The isolate was obtained from an animal initially negative by MAT. When the test was repeated using the isolated strain as the antigen, a titer of 1:100 was obtained.
Table 2.
Microscopic agglutination test (MAT) evaluation of 44 slaughtered sheep.
| Serogroup | Serovar | No. Sheep |
|---|---|---|
| Seropositive* | 9 | |
| Autumnalis | Autumnalis | 2 |
| Bataviae | Bataviae | 2 |
| Sejroe | Hardjo | 2 |
| Icterohaemorrhagiae | Icterohaemorrhagiae | 1 |
| Icterohaemorrhagiae | Lai | 1 |
| Javanica | Poi | 1 |
| Seronegative | 35 |
Seropositive sheep were defined as those whose serum samples had a MAT titer ≥1:100
Discussion
In Brazil, the population of sheep is approximately 15 million (IBGE, 2004). Most of the herds are kept together with cattle and horses and thus, a sheep infected with leptospirosis may be a potential disseminator of the disease to other animal species as well as to humans. In this study isolation of L. noguchii, serogroup Autumnalis from an apparently healthy sheep was obtained. The sequencing of the 16S rRNA gene allowed the classification of this strain as belonging to the L. noguchii species. This is the first report of isolation of L. noguchii from sheep. This pathogenic leptospiral species had previously been isolated only from human, armadillo, toad, spiny rat, opossum, nutria, Mustela nivalis, cattle and Bombina orientalis (Faine et al., 1999).
The existence of sheep carrying L. noguchii may implicate this animal species as playing an important role in the epidemiology of leptospirosis, acting as a maintenance host for infection of cattle and horses by serogroup Autumnalis. Although L. noguchii is a pathogenic species, virulence of the Caco strain could not be demonstrated. The bacterium was obtained from an apparently healthy animal, and experimentally inoculated hamster failed to demonstrate signs of leptospirosis. Possible explanations for this fact include: (i) hamster may not be susceptible to this strain; (ii) the bacteria may have undergone mutations during in vitro growth, resulting in a non-virulent strain (Haake et al., 1991). Within L. noguchii, the only strain that belongs to serogroup Autumnalis is the Fort Bragg strain, a human isolate from USA. Caco strain has not been characterized at serovar level, however it is conceivable that it is different from the serovars described so far.
Serological evidence of leptospiral infection in sheep was found in 20.5% of the animals tested and they reacted against serogroups Autumnalis, Icterohaemorrhagiae, Sejroe, Javanica and Bataviae. A previous study carried out in Brazil by Santa Rosa and Castro (1963) reported reactions predominantly against the Autumnalis leptospiral antigens. More recently, Favero et al. (2002) reported a 0.7% (n=284) seropositive rate among sheep, with reactions against Icterohaemorrhagiae, Butembo, Castellonis, and Hebdomadis antigens. In another study, a seropositive rate of 34.2% (n=1,360) was reported, with predominant reactions occurring with Hardjo (30%), Sentot (16.8%), and Autumnalis (6.3%) serogroups (Herrmann et al., 2004). The etiological significance of these serological findings has not been established.
Considering the economic importance of sheep-raising, the isolation of L. noguchii serogroup Autumnalis strain Caco provides useful information for future epidemiological and serological investigation. Further studies should be carried out to confirm the role of sheep as reservoir of pathogenic leptospires and its implication for public health.
Acknowledgments
This work was supported by CAPES Foundation (Brazilian Government), Bio-Manguinhos, Oswaldo Cruz Foundation, Brazilian Ministry of Health (09224-7), the Brazilian National Research Council (300.861/96-6), and the National Institutes of Health (AI-052473, TW-00919).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azevedo SS, Alves CJ, Andrade JSL, Santos JA, Freitas TD, Batista CSA. Isolation of Leptospira spp. from kidneys of sheep at slaughter. Arq Inst Biol. 2004;71:383–385. [Google Scholar]
- Bahaman AR, Marshall RB, Blackmore DK, Hathaway SW. Isolation of Leptospira interrogans serovar hardjo from sheep in New Zealand. N Z Vet J. 1980;28:1980. doi: 10.1080/00480169.1980.34738. [DOI] [PubMed] [Google Scholar]
- Ciceroni L, Bartoloni A, Pinto A, Guglielmetti P, Valdez Vasquez C, Gamboa Barahona H, Roselli M, Giannico F, Paradisi F. Serological survey of leptospiral infections in sheep, goats and dogs in Cordillera province, Bolivia. New Microbiol. 1997;20:77–81. [PubMed] [Google Scholar]
- Ciceroni L, Lombardo D, Pinto A, Ciarrocchi S, Simeoni J. Prevalence of antibodies to Leptospira serovars in sheep and goats in Alto Adige-South Tyrol. J Vet Med B. 2000;47:217–223. doi: 10.1046/j.1439-0450.2000.00333.x. [DOI] [PubMed] [Google Scholar]
- Cousins DV, Ellis TM, Parkinson J, McGlashan CH. Evidence for sheep as a maintenance host for Leptospira interrogans serovar hardjo. Vet Rec. 1989;124:123–124. doi: 10.1136/vr.124.5.123. [DOI] [PubMed] [Google Scholar]
- Ellis WA, Bryson DG, Neill SD, McParland PJ, Malone FE. Possible involvement of leptospires in abortion, stillbirths and neonatal deaths in sheep. Vet Rec. 1983;112:291–293. doi: 10.1136/vr.112.13.291. [DOI] [PubMed] [Google Scholar]
- Faine S. Guidelines for the control of leptospirosis. World Health Organization; Geneva, Switzerland: 1982. p. 171. [Google Scholar]
- Faine S, Adler B, Bolin C, Perolat P. Leptospira and leptospirosis. Medical Science; Melbourne, Australia: 1999. p. 272. [Google Scholar]
- Favero ACM, Pinheiro SR, Vasconcellos SA, Morais ZM, Ferreira F, Ferreira Neto JS. Most frequent serovars of leptospires in serological tests of Buffaloes, Sheeps, Goats, Horses, Swines and dogs from several Brazilian states. Ciência Rural. 2002;32:613–619. [Google Scholar]
- Gerritsen MJ, Koopmans MJ, Peterse D, Olyhoek T. Sheep as maintenance host for Leptospira interrogans serovar hardjo subtype hardjobovis. Am J Vet Res. 1994;55:1232–1237. [PubMed] [Google Scholar]
- Haake DA, Walker EM, Blanco DR, Bolin CA, Miller MN, Lovett MA. Changes in the surface of Leptospira interrogans serovar Grippotyphosa during in vitro cultivation. Infect Immun. 1991;59:1131–1140. doi: 10.1128/iai.59.3.1131-1140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathaway SC, Little TW, Stevens AE. Serological survey of leptospiral antibodies in sheep from England and Wales. Vet Rec. 1982;110:99–101. doi: 10.1136/vr.110.5.99. [DOI] [PubMed] [Google Scholar]
- Herrmann GP, Lage AP, Moreira EC, Haddad JPA, Resende JR, Rodrigues RO, Leite RL. Seroprevalence of agglutinins anti-Leptospira spp. in sheep from the Southeast and Southwest mesoregions of the State of Rio Grande do Sul, Brazil. Ciência Rural. 2004;34:443–448. [Google Scholar]
- IBGE - Instituto Brasileiro de Geografia e Estatística. [Last access in 09/22/2006];2004 ( http://www.sidra.ibge.gov.br/bda/pecua)
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Natarajaseenivasan K, Ratnam S. Isolation of Leptospira Javanica from sheep. Indian J Anim Sci. 1999;69:759–761. [Google Scholar]
- Santa Rosa CA, Castro AFP. Presença de aglutininas anti-leptospira em soros de ovinos e caprinos no Estado de São Paulo. Arq Inst Biol. 1963;30:93–98. [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. WHO Library Cataloguing-in-Publication Data. 2003. Human leptospirosis: guidance for diagnosis, surveillance and control. [Google Scholar]
- Zamora J, Riedemann S, Tadich N. A serological survey of leptospirosis in sheep in Chile. Rev Latinoam Microbiol. 1999;41:73–76. [PubMed] [Google Scholar]


