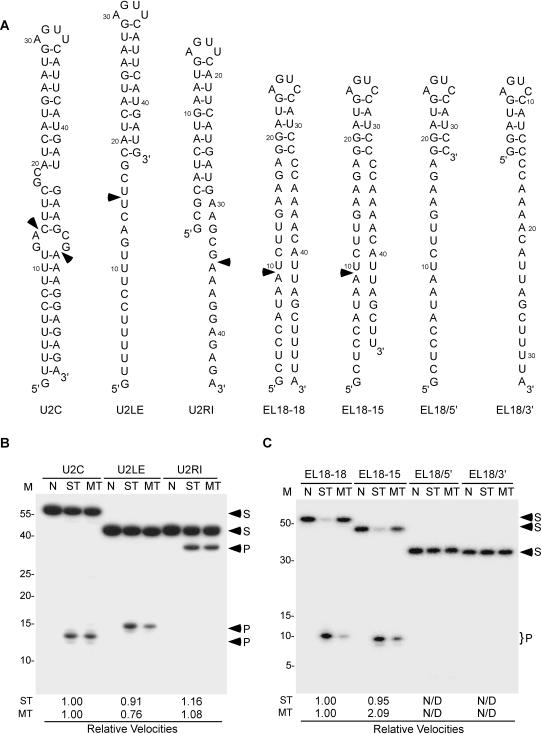
Figure 1. Rnt1p does not require a complete A-form helix for cleavage.
(A) Schematic representations of Rnt1p substrates used in B and C. U2C, U2LE, and U2RI were derived from Rnt1p cleavage site at the 3′-end of U2 snRNA[9]. EL18-18, EL18-15, EL18/5′, and EL18/3′ are derived from the cleavage site at the 3′ end of U5 snRNA[18]. The arrowheads indicate major Rnt1p cleavage sites. (B) and (C) The different 5′-end labeled substrates were incubated in the absence (N) or presence of recombinant Rnt1p. Cleavage was carried out either in enzyme excess to measure the single turnover rate (ST) or in RNA excess to measure the multiple turnover rate (MT). The cleavage products were fractionated by 20% denaturing PAGE and visualized by autoradiogram. The cleavage efficiencies are presented as fractional velocities relative to the parental substrate. The values reflect the average of three independent experiments. The RNA marker (M) is indicated on the left. The positions of the cleavage products (P) and the substrates (S) are indicated by arrowheads on the right.