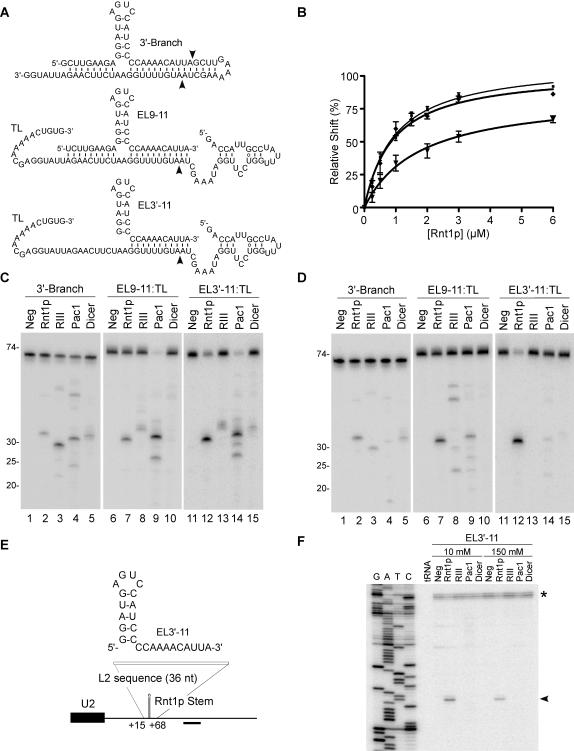
Figure 3. Comparison between inter- and intra-molecular RNA cleavage by different RNase IIIs.
(A) Illustration of the different substrates used in C and D. 3′-Branch indicates a substrate allowing intramolecular cleavage by Rnt1p. EL3′-11 and EL9-11 indicate respectively a guide RNA with a single or two target complementary extensions. The target is indicated by TL. The arrowheads indicate the position of the observed cleavage by Rnt1p. (B) Quantitative analysis of RNA binding to Rnt1p. Increasing concentrations of Rnt1p (0.25 to 6 µM) were incubated with 3 fmol of 3′-Branch (▪), EL9-11:TL (σ) and EL3′-11:TL (τ) and the binding percentage (%) was plotted against the protein concentration. The curve fits were obtained using the Graph Pad Prism 4.0 program. Each data point is an average of four experiments. The target RNA in the trans reactions and the cis RNA were 5′-end labeled and incubated with members of the RNase III family. Rnt1p, bacterial RNase III (RIII), Pac1 and human Dicer were incubated in RNA excess under a 10 mM (C) or 150 mM (D) KCl. The position of the RNA ladder is shown on the left. (E) Sketch of a 36 nt fragment containing sequences complementary to EL3′-11 inserted into a U2 3′-end flanking region to replace a canonical Rnt1p substrate. The position of the oligonucleotide used for primer extension is indicated. (F) Mapping the cleavage of the U2 3′-end region with RNase IIIs. Yeast total RNA (20µg) from YHM111-U2L2 was incubated with EL3′-11 and RNase IIIs in 10 and 150 mM KCl. A primer complementary to the 3′-flanking sequence of U2 snRNA was extended in all cleavage reactions. The reference DNA sequence is shown on the left. The arrowhead indicates a specific cleavage product. The asterisk indicates a secondary structure at the mature U2 3′-end.