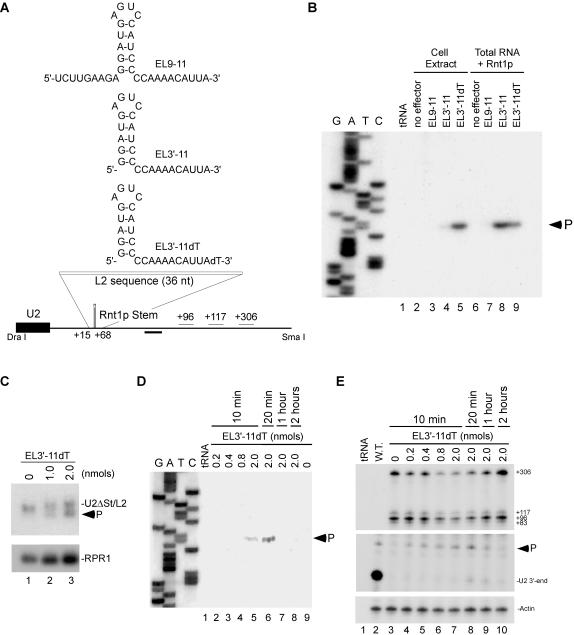
Figure 4. Guide RNA restored cleavage to a mutated Rnt1p cleavage site in vivo.
(A) Secondary structure of RNA guides complementary to a mutated Rnt1p cleavage site at the 3′-end of U2 snRNA (L2). The position of the oligonucleotide used for primer extension is indicated below as well as putative poly(A) signals (+96, +117, and +306). (B) RNA guides were incubated in yeast extract or with yeast total RNA and recombinant Rnt1p for 20 min. The cleavage site was mapped using primer complementary to the 3′-flanking sequence of U2 snRNA. The reference DNA sequence produced using the same primer is shown on the left. The product corresponding to the cleaved RNA is indicated. Bacterial tRNA was used as negative control for the primer extension. (C) Yeast strain YHM111-U2L2 was electroporated with EL3′-11dT and the RNA extracted after 10 minutes of incubation. The RNA bands were analyzed by northern blot using a probe complementary to mature U2 snRNA sequence. A probe directed against RPR1 was used as loading control. The arrowhead indicates the position of the cleavage product. (D) Cleavage site mapping of yeast YHM111-U2L2 electroporated with EL3′-11dT. Total RNA was extracted between 10 minutes and 2 hours post-electroporation and annealed to the primer used in B. The reference DNA sequence is shown on the left. The product corresponding to the cleaved RNA is indicated. (E) Analysis of U2 snRNA 3′-end formation. RNA samples described in D were hybridized to an RNA probe (DraI-SmaI fragment) complementary to the 3′- flanking sequences of U2 snRNA, and digested with RNase T1. The mature U2 3′-end and the ends of the extended forms are indicated on the right. The Rnt1p-directed cleavage product is indicated by an arrowhead. The position of the different 3′-ends detected is indicated using wild-type U2 sequence as reference. A probe against actin was used as internal control for loading and quantification.