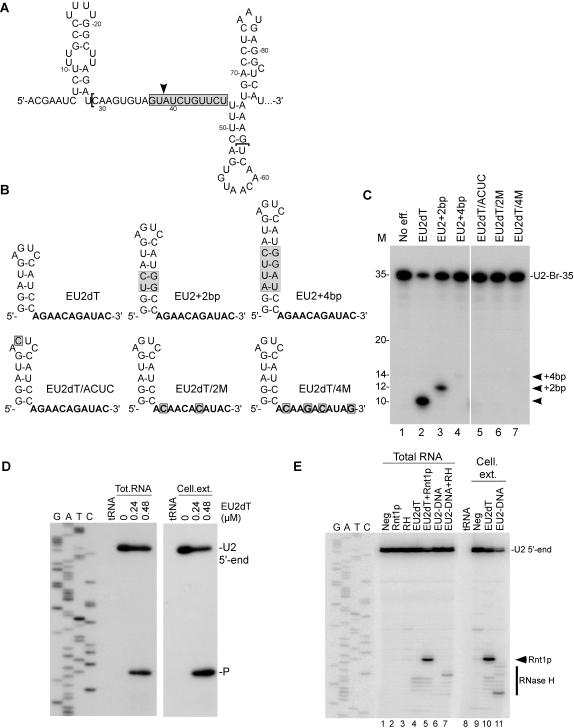
Figure 5. Guide RNA directs sequence specific cleavage in a natural RNA sequence.
(A) Secondary structure of the U2 snRNA branch site region (nucleotides 1 to 86). The gray box, the arrowhead and the brackets represent respectively the targeted region by Rnt1p, the anticipated cleavage site by Rnt1p and the region used for in vitro cleavage assays (U2-Br-35). (B) Sketches representing the secondary structure of guides recognized by Rnt1p and complementarity to the U2 branch site. Sequences in bold represent the nucleotides complementary to the U2 target. The gray boxes indicate mutations relative to the control (EU2dT). (C) In vitro cleavage of 5′-end labeled U2-Br-35 with Rnt1p and the different RNA guides. The cleavage reactions were performed in RNA excess with a guide/target ratio of 1:1. The positions of the cleavage products are indicated on the right and the RNA marker is displayed on the left. (D) Total yeast RNA and recombinant Rnt1p or yeast cell extract prepared from strain YHM111-U2L2 were used to analyze Rnt1p-directed cleavage using EU2dT. Primer complementary to the 3′-flanking sequence of the U2 snRNA branch site was extended on the extracted RNA to map the cleavage site. The reference DNA sequence produced using the same primer is shown on the left. The product corresponding to the cleaved RNA and the U2 5′-end are indicated on the right. Bacterial tRNA was used as negative control for the primer extension. (E) Cleavage comparison between Rnt1p and RNase H in total RNA or cell extract prepared from yeast YHM111-U2L2. The cleavage specificity was determined by primer extension. The reference DNA sequence produced using the same primer is shown on the left. The RNA guide and the DNA oligo used with RNase H targeted the same nucleotides. The positions of Rnt1p and RNase H cleavage products and the U2 5′-end are indicated on the right.