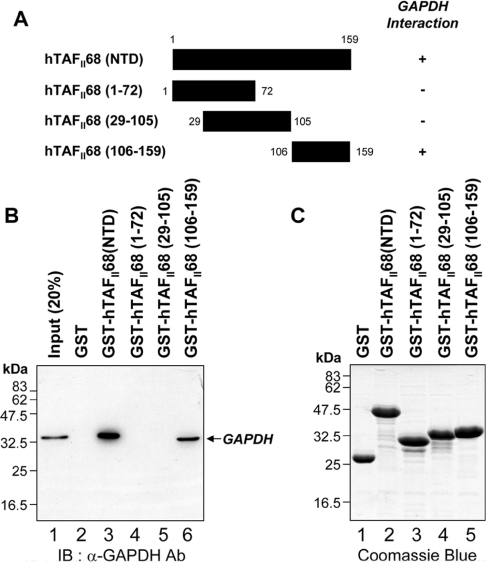
Figure 4. Mapping the hTAFII68 (NTD) region that interacts with GAPDH.
(A) Schematic representation of the GST–hTAFII68 (NTD) fusion proteins and their ability to bind to GAPDH. Numbers refer to amino acid residues, and binding ability is indicated by + or −. (B) Strong binding of GAPDH to GST–hTAFII68 (106–159). Recombinant GST–hTAFII68 (NTD) deletion mutants were incubated with HEK-293T cell lysates. Following GST pull-down assays, the bound proteins were eluted with SDS loading buffer and analysed by Western blotting with an anti-GAPDH antibody. The positions of molecular mass markers and of GAPDH are indicated. Three independent experiments were performed, all of which gave similar results. Lane 1, 20% input; lane 2, GST alone; lane 3, GST–hTAFII68 (NTD); lane 4, GST–hTAFII68 (1–72); lane 5, GST–hTAFII68 (29–105); lane 6, GST–hTAFII68 (106–159). IB, immunoblotting; Ab, antibody. (C) Coomassie Blue staining of the GST–hTAFII68 deletions. The amounts of GST-fusion proteins utilized in these assays were fractionated on SDS/PAGE (15% gels) and visualized by Coomassie Blue staining. Lane 1, GST alone; lane 2, GST–hTAFII68 (NTD); lane 3, GST–hTAFII68 (1–72); lane 4, GST–hTAFII68 (29–105); lane 5, GST–hTAFII68 (106–159).