Abstract
Functional and structural properties of several truncated or mutated variants of Candida albicans Gfa1p (glucosamine-6-phosphate synthase) were compared with those of the wild-type enzyme. Fragments encompassing residues 1–345 and 346–712 of Gfa1p, expressed heterogeneously in bacterial host as His6 fusions, were identified as the functional GAH (glutamine amidehydrolysing) and ISOM (hexose phosphate-isomerizing) domains respectively. It was found that the native GAH domain is monomeric, whereas the native ISOM domain forms tetramers, as does the whole enzyme. Spectrofluorimetric and kinetic studies of the isolated domains, the Δ218–283Gfa1p mutein and the wild-type enzyme revealed that the binding site for the feedback inhibitor, uridine 5′-diphospho-N-acetyl-D-glucosamine, is located in the ISOM domain. Inhibitor binding affects amidohydrolysing activity of the GAH domain and, as a consequence, the GlcN-6-P (D-glucosamine-6-phosphate)-synthetic activity of the whole enzyme. The fragment containing residues 218–283 is neither involved in ligand binding nor in protein oligomerization. Comparison of the catalytic activities of Gfa1pV711F, Δ709–712Gfa1p, Gfa1pW97F and Gfa1pW97G with those of the native Gfa1p and the isolated domains provided evidence for an intramolecular channel connecting the GAH and ISOM domains of Gfa1p. The channel becomes leaky upon deletion of amino acids 709–712 and in the W97F and W97G mutants. The Trp97 residue was found to function as a molecular gate, opening and closing the channel. The W97G and V711F mutations resulted in an almost complete elimination of the GlcN-6-P-synthetic activity, with the retention of the amidohydrolase and sugar phosphate-isomerizing activities.
Keywords: amidotransferase, glucosamine-6-phosphate synthase (Gfa1p), glutamine amide hydrolysing (GAH) domain, hexosephosphate-isomerizing (ISOM) domain, site-directed mutagenesis, intramolecular channel
Abbreviations: CPS, carbamoyl phosphate synthetase; DTT, dithiothreitol; Fru-6-P, D-fructose-6-phosphate; GAH, glutamine amide hydrolysing; GAHp–His6, His-tagged C. albicans GAH domain; Gfa1p, glucosamine-6-phosphate synthase; GlcN-6-P, D-glucosamine-6-phosphate; Gln, glutamine; Glu, glutamate; GLUPA, γ-glutamyl-p-nitroanilide; His6–ISOMp, His-tagged C. albicans ISOM domain; IDA, ion-dependent adhesion; ISOM, hexosephosphate-isomerizing; LB, Luria–Bertani; PKA, protein kinase A; UDP-GlcNAc, uridine 5′-diphospho-N-acetyl-D-glucosamine
INTRODUCTION
L-Glutamine:D-fructose-6-phosphate amidotransferase (EC 2.6.1.16) is an ubiquitous enzyme, catalysing the first committed step in the amino sugar biosynthetic pathway in prokaryotic and eukaryotic organisms. The enzyme catalyses a complex reaction: L-Gln+D-fructose-6-phosphate→D-glucosamine-6-phosphate+L-Glu (where L-Gln is glutamine and L-Glu is glutamate), that involves ammonia transfer and sugar phosphate isomerization. GlcN-6-P (D-glucosamine-6-phosphate) synthase is an important point of metabolic control of amino sugar biosynthesis, the mammalian enzyme is proposed to be involved in the phenomenon of hexosamine-induced insulin resistance in diabetes [1] and the fungal GlcN-6-P synthase has been proposed as a potential target in antifungal chemotherapy [2]. For all these reasons, the enzyme, especially its eukaryotic type, is an interesting subject of study. The prokaryotic GlcN-6-P synthase has been relatively well characterized. The Escherichia coli enzyme has been purified to homogeneity [3], details of the molecular mechanism of catalysis have been deduced ([4] and references cited therein) and its three-dimensional structure has been determined [5]. Previous studies on E. coli GlcN-6-P synthase revealed that the enzyme is a homodimer. Each subunit is composed of two domains that can be released from the native protein upon limited proteolysis with chymotrypsin: the N-terminal GAH (glutamine amidohydrolase; residues 1–239) and the C-terminal ISOM [hexosephosphate-isomerizing (ketose/aldose isomerase); residues 240–608] domains [5]. The former binds L-Gln and hydrolyses it to Glu and ammonia. The latter utilizes the released ammonia for the conversion of D-fructose-6-phosphate into D-glucosamine-6-phosphate. Such a modular structure is characteristic for all amidotransferases composed of an amidohydrolase domain fused with an acceptor domain [6]. It is assumed that ammonia generated at the amidohydrolase domain is transferred to the second domain through the intramolecular channel. In most amidotransferases, the channel is accessible for exogenous ammonia; however, the GlcN-6-P synthase seems to be unique in its apparent inability to use this substrate [6]. The presence of a solvent-inaccessible relatively long channel composed of several hydrophobic amino acid residues was demonstrated in the crystal structure of E. coli GlcN-6-P synthase [5]. However, some evidence was presented for the close proximity of the domains' active sites [7] and, recently, for a hinged movement of the amidohydrolase domain towards the isomerase domain, induced upon substrate binding [8].
Only limited structural data are available for the eukaryotic GlcN-6-P synthase. There is little doubt that the essential active site amino acid residues are highly conserved among GlcN-6-P synthases. However, the eukaryotic enzyme differs from its prokaryotic counterpart in a number of features. The most important of these is the quaternary structure and regulation of the enzymatic activity. The eukaryotic GlcN-6-P synthase is undoubtedly homotetrameric, its activity is regulated by a feedback inhibition by UDP-GlcNAc (uridine 5′-diphospho-N-acetyl-D-glucosamine), as well as by phosphorylation/dephosphorylation mediated by PKA (protein kinase A) and protein phosphatase [9]. Moreover, the results of multiple sequence alignment analysis of the enzyme revealed the presence of a 30–80 residue fragment of unknown function at the C-terminal part of the eukaryotic GAH domain. No such fragment was observed in any known prokaryotic versions of the enzyme [9].
In the present study, we describe the results of our studies on GlcN-6-P synthase from the human pathogenic fungus Candida albicans (Gfa1p), involving structural and functional characterization of several truncated or mutated variants of this enzyme.
MATERIALS AND METHODS
Bacterial strains and media
The E. coli XL1-Blue strain from Stratagene was used in all cloning procedures. The E. coli BL21(DE3) pLysS strain from Novagen was used for the overexpression of both domains of C. albicans GlcN-6-P synthase. E. coli strains were cultured at 37 °C on LB (Luria–Bertani) solid medium [1x.0% (w/v) NaCl, 1.0% (w/v) tryptone, 0.5% yeast extract and 1.5% (w/v) agar] and LB liquid medium [1.0% (w/v) NaCl, 1.0% (w/v) tryptone and 0.5% yeast extract] supplemented with 0.1 mg/ml ampicillin when necessary.
Plasmids, enzymes and other materials
The plasmids used were: pET23b (Novagen), pUC19 (Fermentas), pUC19-CaGFA1 (C. albicans GFA1) and pET23b-CaGFA1 [10]. Restriction and modification enzymes were purchased from Fermentas and New England Biolabs. Purification of His-tagged proteins was performed on Ni2+-IDA (ion-dependent adhesion) agarose (His·Bind Resin, Novagen). DNA molecular mass markers were from DNA-Gdańsk II s.c., Poland. The GelCode™ Phosphoprotein Staining Kit was from Pierce. Yeast Glc-6-P dehydrogenase and other reagents were from Sigma.
DNA manipulations
Isolation of plasmid DNA was carried out according to the protocol of the Plasmid Mini kit (A&A Biotechnology). DNA fragments were isolated from agarose gels following the standard procedure of the DNA Gel-Out kit (A&A Biotechnology). DNA purification after enzyme treatment was performed according to the instructions in the DNA Clean-up kit (A&A Biotechnology). DNA digestion with restriction enzymes was carried out according to the enzyme supplier's instructions. DNA fragments were ligated and E. coli cells were transformed according to the standard methods [11].
Construction of expression plasmids containing domain-coding sequences
The fragment of the GFA1 gene encoding residues 1–345 of Gfa1p was amplified from the pUC19-CaGFA1 plasmid by PCR. The primers used in the amplification were: Gln1-KN (Table 1) and Gln2-HX (5′-cctgccaagcttctcgagaatttcattcaattccatttctaaagtttg-3′). The artificial cloning sites KpnI and NdeI (Gln1-KN), and XhoI and HindIII (Gln2-HX), at the 5′-ends of the primers were introduced to facilitate the cloning procedure. The PCR product (1071 bp) was purified from an agarose gel and was cloned directionally in between the KpnI and HindIII sites of the pUC19 vector and then re-cloned into the pET23b plasmid, giving a recombinant expression plasmid pET23b-GLN (4628 bp). The identity of the plasmids was confirmed by restriction analysis and DNA sequencing. The obtained construct encoded the putative GAH domain of Gfa1p, containing an additional octapeptide (Lys-Glu-His-His-His-His-His-His) at the C-terminus (GAHp–His6).
Table 1. PCR primers used for the construction of site-directed mutants of GFA1.
Restriction recognition sites are underlined. Capital letters indicate mismatches with the original GFA1 sequence (bold, introducing mutation; normal, introducing restriction recognition site).
| Mutation | Primer name | Primer sequence |
|---|---|---|
| None | Gln1-KN | 5′-cctggcggtacccatatgtgtggtatttttggttacg-3′ |
| cgfa-bam | 5′-cacgggatccttactcaacagtaactgatttagcca-3′ | |
| W97G | Trpgly-3 | 5′-catgtgtGgCgcCtctagtatgagcaatacc-3′ |
| Trpgly-5 | 5′-catactagaGgcGcCacacatggtcaacc-3′ | |
| W97F | Trpphe-3 | 5′-agcGAaAcGCgtatgagcaataccaacatg-3′ |
| Trpphe-5 | 5′-ctcatacGCgTtTCgctacacatggtcaac-3′ | |
| V711F | Valphe | 5′-cctgccctcgagttactcaaAagtaactgatttagccaag-3′ |
Analogously, the respective GFA1 gene fragment encoding residues 356–712 of Gfa1p was amplified from the pUC19-CaGFA1 plasmid by PCR using the following primers: Fru1-BNK (5′-cctggcggatcccatatgcaccaccaccaccaccacggtaccatgaaaggcccctataaacattttatg-3′) and Fru2-Eco (5′-cctgccgaattcttactcaacagtaactgatttagcca-3′). The artificial cloning sites BamHI and NdeI (Fru1-BNK) and EcoRI (Fru2-Eco), at the 5′-ends of primers (underlined) were introduced to facilitate the cloning procedure. After cloning into pUC19 and subsequent re-cloning into pET23b, a recombinant expression plasmid pET23b-FRU (4755 bp) was obtained. The construct encoded the putative ISOM domain of C. albicans Gfa1p containing an additional nonapeptide (Met-His-His-His-His-His-His-Gly-Thr) at the N-terminus (His6–ISOMp).
Construction of expression plasmids containing truncated versions of GFA1
Δ218–283GFA1
Two fragments were amplified from the pUC19-CaGFA1 template by PCR using the following primers: fragment 1, GLN1-KN (Table 1) and Ingln-X (5′-tccaggctcgagatcagttttaacaccaaccaataatg-3′); fragment 2, Infru-X (5′-cctaccctcgagttctttttatcttctgatcctgc-3′) and Fru2-Eco (see above). Artificial restriction recognition sites for XhoI are underlined. The PCR products were purified and digested with Bsu15I and XhoI for the GLN1-KN/Ingln-X product, and XhoI and EcoRI for the Infru-X/Fru2-Eco product. This gave three fragments of different sizes in each case (digests 1 and 2). A 413 bp fragment was isolated from digest 1 and a 1293 bp fragment from digest 2. The pET23b-CaGFA plasmid was digested with Bsu15I and EcoRI and a 3865 bp fragment was isolated from the digest. The 413 bp, 1293 bp and 3865 bp fragments were ligated together and E. coli XL1-Blue cells were transformed with the post-ligation mixture. The resulting plasmid construct pET23b-CaGFA1Δ218–283 was isolated and sequenced to confirm that it encodes the truncated Gfa1p lacking residues 218–283.
Δ709–712GFA1
A GFA1 fragment was amplified from the pUC19-CaGFA1 template by PCR, using the following primers: Trpgly-5 (Table 1) and GFA709X (5′-atgacgctcgagttatgatttagccaagttacgagg-3′). The PCR product was purified and digested with Bsp119I and XhoI to give three products, of which the 1052 bp one was isolated. The pET23b-CaGFA plasmid was digested with the same restriction enzymes and a 4666 bp fragment was isolated from the digest. The isolated fragments were ligated, and E. coli XL1-Blue cells were transformed with the ligation product. The resulting plasmid construct pET23b-CaGFA1Δ709–712 was isolated and its identity was confirmed by sequencing.
Construction of expression plasmids containing site-directed mutant versions of GFA1
The site-directed mutants W97G and W97F of Gfa1p were constructed using the overlap extension method with pUC19-CaGFA1 as the template. Primers used for PCR are listed in Table 1. The V711F mutant was amplified from pUC19-CaGFA1 with Gln1-KN and Valphe primers. The PCR products were isolated, digested with MluI, KpnI and BglII (W97F), KpnI, KasI and BglII (W97G), and Bsp119I and XhoI (V711F), and ligated into the pET23b-CaGFA1 vector, digested with the appropriate restriction enzymes. E. coli XL1-Blue cells were transformed with the ligation products. The resulting plasmid constructs pET23b-CaGFA1W97G, pET23b-CaGFA1W97F and pET23b-CaGFA1V711F were isolated and their identity was confirmed by sequencing. Construction of the pET23b-CaGFA1S208A plasmid was described previously [12].
Protein expression and purification
E. coli BL21(DE3)pLysS cells, transformed with the appropriate expression plasmid, were grown in LB liquid medium at 37 °C. Expression was induced by the addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside to the cultures grown to D600=0.6–0.8 and incubation was continued for a further 5 h. Cells were harvested by centrifugation at 5000 g for 10 min at 4 °C.
The His-tagged putative domains of Gfa1p were purified by metal-affinity chromatography. The bacterial pellet was suspended in buffer A (20 mM Tris/HCl, pH 7.9, 500 mM NaCl, 5 mM imidazole, 0.1% Triton X-100 and 0.5 mM PMSF) and the cells were disrupted by sonication (5×30 s bursts with 30 s intervals at a power setting of 30 W using a Branson sonifier 250) on ice. The total lysate was centrifuged at 16000 g for 20 min at 4 °C. The supernatant (crude extract) was applied to a Ni2+-IDA agarose (His·Bind Resin) column, bed volume 10 ml, which was pre-equilibrated with 4 vol of buffer A. Next, the column was washed with 20 ml of the same buffer, followed by washing with 40 ml of buffer W (20 mM Tris/HCl, pH 7.9, 500 mM NaCl, 80 mM imidazole, 0.1% Triton X-100 and 0.5 mM PMSF). The His-tagged proteins were eluted by two 5 ml portions of elution buffer E (20 mM Tris/HCl, pH 7.9, 500 mM imidazole, 500 mM NaCl, 0.1% Triton X-100 and 0.5 mM PMSF). For further assays, the eluates were concentrated by ultrafiltration using Vivaspin concentrators (10 kDa cut-off limit; Viva Science Ltd.) at 7000 g for 30 min at 4 °C and the buffer was exchanged for buffer B [25 mM potassium phosphate, pH 7.5, 1 mM EDTA, 1 mM DTT (dithiothreitol) and 0.5 mM PMSF] using the HiTrap™ Desalting Columns (Pharmacia LKB).
The wild-type Gfa1p and the truncated and site-directed mutant variants were purified by a multi-step procedure, essentially as described previously [10].
Determination of enzymatic activities and enzyme kinetic analysis
The amidohydrolysing activity was determined using GLUPA (L-γ-glutamyl-p-nitroanilide) as a substrate. Mixtures containing 1 mM GLUPA, 1 mM EDTA, 1 mM DTT and the appropriately diluted enzyme preparation in 20 mM Hepes buffer (pH 7.5) were incubated at 25 °C. In some experiments, 10 mM Fru-6-P (D-fructose-6-phosphate) was also included as a component of the incubation mixture. The released p-nitroaniline was quantified spectrophotometrically at 420 nm, assuming ϵ420(p-nitroaniline)=9300 M−1 cm−1. One unit of specific activity was defined as the amount of the enzyme that catalysed formation of 1 μmol of p-nitroaniline/min per mg of protein.
The rate of L-Gln hydrolysis was determined by quantification of the released ammonia with the Nessler's reagent. Mixtures containing 10 mM L-Gln, 1 mM EDTA, 1 mM DTT and an appropriately diluted enzyme preparation in 20 mM Hepes buffer (pH 7.5) were incubated at 25 °C. Aliquots (0.1 ml) were withdrawn at 5 min intervals, diluted with 1.4 ml of water and then 0.2 ml of a commercial solution of the Nessler's reagent was added. The mixtures were left for 10 min at room temperature (20 °C) and absorbance at 480 nm was measured. The released ammonia was quantified, assuming a molecular absorbance index for the yellow product of the Nessler reaction with a standard NH4Cl solution, ϵ480=3×108 M−1 cm−1.
The ISOM activity was determined using the Glc-6-P dehydrogenase-coupled assay, as described by Noltman et al. [13]. Incubation mixtures containing 2 mM Fru-6-P, 0.5 mM NADP+, 1 mM EDTA, 2.6 units/ml Glc-6-P dehydrogenase and appropriately diluted GlcN-6-P synthase preparation in 50 mM Tris/HCl buffer (pH 7.5) were incubated at 25 °C. The reduction of NADP+ to NADPH was monitored spectrophotometrically at 340 nm, assuming ϵ340(NADPH)=6220 M−1cm−1. One unit of specific activity was defined as the amount of the enzyme that catalysed formation of 1 μmol of Glc-6-P/min per mg of protein. GlcN-6-P-synthetic activity was assayed as described previously [14].
For the determination of kinetic constants, the amidohydrolysing, Fru-6-P-isomerizing or GlcN-6-P-synthetic activities were determined at variable (0–10 mM) initial concentrations of appropriate substrates. The data were plotted as Michaelis–Menten graphs. The kinetic data were subjected to the non-linear regression analysis using the Enzyme Kinetics Module 1.10 for Sigma Plot software. The Km and kcat values were determined in triplicate, to give the means±S.D.
Phosphorylation of Gfa1p, Gfa1pS208A and GAHp-His6 in vitro
Gfa1p, Gfa1pS208A or GAHp–His6 (100 μg/ml each) were incubated for 30 min at 25 °C with 10 μM cAMP, 1 mM ATP, 10 mM EDTA, 40 mM NaF and, if necessary, with 10 μg/ml of the specific PKA inhibitor, H-89. The reaction was initiated by the addition of the cAMP-dependent kinase from bovine heart (30 units/ml). After 30 min, H-89 was added to each sample to stop the PKA activity. Samples were subjected to SDS/PAGE analysis and assayed for amidohydrolysing or GlcN-6-P-synthetic activity.
Detection of phosphorylated proteins in SDS/PAGE gels
The gels obtained after SDS/PAGE separation of protein mixtures incubated with PKA, were stained with the GelCode™ Phosphoprotein Staining kit, according to the protocol provided by the manufacturer. Gel images were documented, then the gels were destained by several washings with demineralized water and stained with Coomassie Brilliant Blue R-250 to visualize all the protein bands.
Steady-state fluorescence measurements
Fluorescence emission spectra were obtained using the Perkin Elmer LS55 spectrofluorimeter at room temperature. The excitation wavelength was set at 300 nm for selective excitation of tryptophan fluorescence. Incubation mixtures contained 0.3 μM of the respective domain, mutant or wild-type Gfa1p, 1 mM EDTA and/or a ligand at appropriate concentration in 25 mM phosphate buffer (pH 7.5). The incubation solution was stirred continuously in a 1.0 cm×1.0 cm cuvette. The bandwidths for both the excitation and the emission monochromators were 4 nm. The spectra were corrected for background and Raman scattering by subtracting buffer spectra containing all ingredients except the protein.
Molecular-mass determination of the native protein
Size-exclusion chromatography was performed on Superdex 200 HR 10/30 (Pharmacia LKB), eluted at 0.5 ml/min by 25 mM potassium phosphate buffer (pH 7.0) containing 0.15 M NaCl, 1 mM EDTA and 1 mM DTT. Protein elution was monitored at 280 nm. The molecular-mass standards were: thyroglobulin (669 kDa), apoferritin (443 kDa), α-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa) and carbonic anhydrase (29 kDa).
Other methods
The wild-type Gfa1p was overexpressed in E. coli and purified to near homogeneity according to the multi-step procedure described by Sachadyn et al. [10]. Construction and purification of His6–Gfa1p and Gfa1p–His6 were as described previously [15]. Protein concentration was determined by the Bradford method [16]. Discontinuous SDS/PAGE was performed by the method of Laemmli [17] using a 10% (w/v) separating gel and a 5% (w/v) stacking gel.
RESULTS
Structural alignment of bacterial and fungal Gfa1p sequences
Amino acid sequences of E. coli and C. albicans GlcN-6-P synthases were aligned and the result is shown in Figure 1. The sequences demonstrate 36% identity and 52% similarity; however, several gaps, indicating regions which are absent from the shorter bacterial variant of the enzyme, are clearly shown. Most of them are present in the N-terminal (residues 1–239) part of the bacterial protein, constituting the GAH domain, with the largest one corresponding to residues 218–283 of the C. albicans enzyme.
Figure 1. Alignment of amino acid sequences of C. albicans and E. coli GlcN-6-P synthases.
The linker between the GAH and ISOM domains of E. coli GlcN-6-P synthase and the residues substituted in the recombinant versions of the C. albicans enzyme, constructed for the purpose of the present study, are indicated. Numbering for the C. albicans Gfa1p is shown. White letters on a black background indicate conserved residues. Ca, C. albicans; Ec, E. coli.
Construction, expression, purification and characterization of the His6-tagged domains of C. albicans Gfa1p
The two-domain structure of E. coli GlcN-6-P synthase is well documented, as well as the roles played by each domain in the mechanism of catalysis. As shown in Figure 2, the GAH domain (residues 1–239) is connected to the ISOM (residues 249–608) by a flexible nonapeptide linker. In the first step of the present study, we constructed two separate fragments of the C. albicans Gfa1p and characterized their functional properties. One of them comprised residues 1–345 and the other residues 346–712. Both constructs contained His6 tags to facilitate their purification, however, in the 1–345 construct, the tag was located at the C-terminus, whereas in the 356–712 construct it was at the N-terminus. Such localizations were an obvious consequence of the results of our previous studies on the construction and functional properties of the N- and C-terminally His6-tagged versions of the complete enzyme. We showed that the presence of a His6 tag at either end almost entirely inactivated the GlcN-6-P-synthesizing activity of the enzyme. On the other hand, the N-terminally tagged enzyme lost the ability to hydrolyse L-Gln but not to interconvert Glc-6-P and Fru-6-P, whereas the opposite was found for the C-terminally tagged enzyme [15].
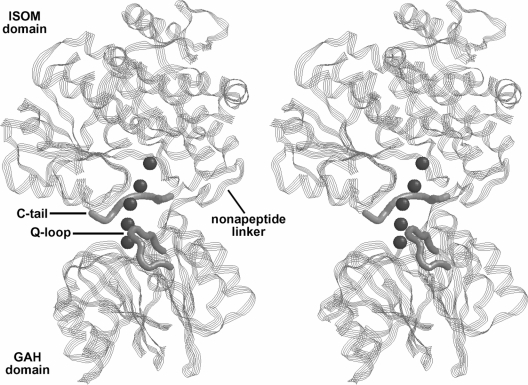
Figure 2. Stereo view of the three dimensional structure of E. coli GlcN-6-P synthase.
The functional domains, some relevant fragments and the intramolecular channel (small spheres) are indicated. The image has been generated from PDB ID 1JXA [5].
Expression plasmids pET23b-GLN and pET23b-FRU were constructed and their identities were confirmed by restriction analysis and DNA sequencing. The fusion proteins, called GAHp–His6 and His6–ISOMp, were overproduced in E. coli BL21(DE3)pLysS cells that had been transformed with the respective overexpression plasmid. Densitometric analysis revealed that, in each case, the recombinant protein constituted 15–20% of the total cytoplasmic protein pool. GAHp–His6 and His6–ISOMp were purified by metal-affinity chromatography using the Ni2+-IDA–agarose resin. A single-step elution resulted in both cases in almost homogeneous protein preparations (approx. 98% homogeneity), isolated with 79−81% yield. The exemplary results of the SDS/PAGE analysis of His6–ISOMp purification are shown in Figure 3.
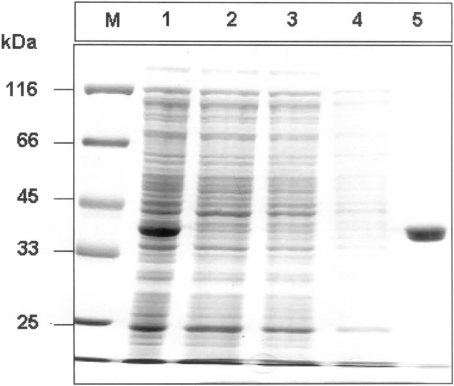
Figure 3. SDS/PAGE analysis of the purification of N-terminally His6-tagged residues 346–712 of Gfa1p overexpressed in E. coli.
Lane M, molecular-mass markers (in kDa); lane 1, total lysate of E. coli BL21(DE3)pLysS pET23b-FRU cells; lane 2, total lysate of control E. coli BL21(DE3)pLysS pET23b cells; lanes 3 and 4, wash fractions with buffer W; and, lane 5, fraction eluted by 0.5 M imidazole.
The purified GAHp–His6 and His6–ISOMp were characterized in terms of their enzymatic activities and molecular mass, and these properties were compared with those of the wild-type complete enzyme and its His6-tagged fusions. The amidohydrolysing activity was tested using GLUPA as the substrate. The ISOM activity was determined with Fru-6-P as the substrate, by continuous monitoring of the Glc-6-P formation in the Glc-6-P dehydrogenase-coupled assay. The GlcN-6-P formation was followed by the modified Elson–Morgan reaction [14]. The obtained results are summarized in Table 2. As expected, both GAHp–His6 and His6–ISOMp were completely devoid of GlcN-6-P-synthetic activity. On the other hand, both fragments exhibited partial activities: GAHp–His6 catalysed GLUPA hydrolysis and His6–ISOMp was able to isomerize Fru-6-P into Glc-6-P. Kinetic parameters of both reactions were very similar to those found for the wild-type intact enzyme. The catalytic properties of GAHp–His6 were also very similar to those of Gfa1p–His6, whereas His6–ISOMp demonstrated catalytic properties almost identical to those of His6–Gfa1p. Therefore, there is little doubt that in C. albicans Gfa1p, the 1–345 fragment constitutes the GAH domain and the remaining part is the ISOM domain.
Table 2. Kinetic parameters of reactions catalysed by Gfa1p, its His6-tagged variants and His6-tagged domains.
The results are the means±S.D. for three independent determinations. ND, not determined.
| Amidohydrolysing activity | ISOM activity | GlcN-6-P-synthetic activity | |||||
|---|---|---|---|---|---|---|---|
| Enzymatic protein | Km(GLUPA) (mM) | kcat (min−1) | Km(Fru-6-P) (mM) | kcat (min−1) | Km(Gln) (mM) | Km(Fru-6-P) (mM) | kcat (min−1) |
| Gfa1p | 0.67±0.04 | 0.74±0.05 | 1.20±0.08 | 1.28±0.09 | 0.34±0.01 | 1.41±0.05 | 720±24 |
| His6–Gfa1p | 12.2±0.9 | 0.055±0.004 | 1.08±0.07 | 1.27±0.06 | ND | ND | ND |
| Gfa1p–His6 | 0.64±0.08 | 0.80±0.07 | 6.50±0.31 | 0.24±0.03 | ND | ND | 12.6±0.4 |
| GAHp–His6 | 0.31±0.02 | 0.45±0.03 | ND | ND | ND | ND | ND |
| His6–ISOMp | ND | ND | 1.40±0.11 | 0.55±0.03 | ND | ND | ND |
The molecular mass of the native GAHp–His6, determined under non-denaturing conditions (size-exclusion chromatography) was 42±2 kDa and that of the denatured form of this protein (determined by SDS/PAGE) was 41±0.5 kDa. The values found for His6–ISOMp were 159±2 kDa and 39.5±0.5 kDa respectively. One may therefore conclude that GAHp–His6 is monomeric in solution, whereas His6–ISOMp is found as a homotetramer.
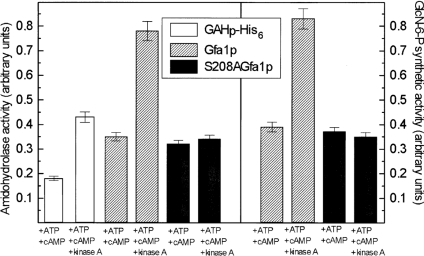
Our previous study showed that C. albicans Gfa1p can be phosphorylated by a cAMP-dependent protein kinase and that the GlcN-6-P-synthetic activity of the enzyme increases upon PKA-driven phosphorylation [14]. Moreover, we identified the Ser208 residue as a unique phosphorylation site [12]. In the present study, we checked whether the isolated GAH domain of Gfa1p, containing Ser208, was a substrate for a commercially available cAMP-dependent protein kinase from bovine heart. GAHp–His6 was incubated with the protein kinase under conditions optimal for protein phosphorylation and its amidohydrolysing activity was monitored. For comparison, the same activity was measured for Gfa1p and Gfa1pS208A, together with the determination of possible changes in the GlcN-6-P-synthetic activity. The results of these experiments are shown in Figure 4. It was found that incubation of GAHp–His6 and Gfa1p with PKA in the presence of ATP and cAMP resulted in an almost 2-fold increase of the amidohydrolysing activity of both proteins, which correlated closely with a very similar enhancement of the synthetic activity of the whole enzyme. On the other hand, no change in both activities of Gfa1pS208A, which lacks the phosphorylation site, was noted. It seems therefore very likely that the enhanced GlcN-6-P-synthetic activity of Gfa1p phosphorylated at Ser208 is a direct consequence of the elevated amidohydrolysing activity of GAH.
Figure 4. Changes in enzymatic activities of GAH-His6, Gfa1p and S208AGfa1p upon phosphorylation with PKA.
Samples of Gfa1p, S208AGfa1p or GAH-His6 were incubated for 30 min in the presence of indicated compounds. A specific kinase A inhibitor, H-89 (10 μg/ml), was then added and the samples were assayed for amidohydrolysing and/or GlcN-6-P-synthetic activity.
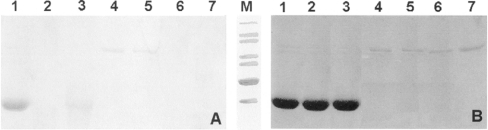
In order to confirm protein phosphorylation and the identity of the phosphorylation site, SDS/PAGE analysis of the reaction mixtures, followed by specific gel staining was performed. The results of this experiment are shown in Figure 5. The gel shown in Figure 5(A) was stained with the GelCode™ kit, allowing a specific detection of the O-phosphorylated serine residues. The stained bands can be clearly seen in lanes 1, 4 and 5 (Figure 5A), thus confirming the presence of an O-phosphorylated serine residue in the PKA-treated GAHp–His6 and Gfa1p. The presence of Fru-6-P in the incubation mixture did not affect Gfa1p phosphorylation. A specific PKA inhibitor, H-89, almost stopped phosphorylation of GAHp–His6 (Figure 5A, note the very faint band in lane 3). No band can be seen in lane 6 (Figure 5A), thus confirming that Ser208 is indeed the residue phosphorylated by PKA in Gfa1p.
Figure 5. SDS/PAGE analysis of GAHp–His6, Gfa1p and Gfa1pS208A phosphorylation by PKA.
Mixtures obtained after a 30 min incubation of a given protein with indicated components were separated by SDS/PAGE. (A) Gel stained with the GelCode™ kit. (B) The same gel as (A) stained with Coomassie Brilliant Blue. Lane 1, GAHp–His6 and PKA; lane 2, GAHp–His6; lane 3, GAHp–His6, PKA and H-89; lane 4, Gfa1p and PKA; lane 5, Gfa1p, PKA and 10 mM Fru-6-P; lane 6, S208AGfa1p and PKA; lane 7, Gfa1p. Each mixture contained 10 μM cAMP, 1 mM ATP, 10 mM EDTA and 40 mM NaF. Protein load: 10 μg (lanes 1–3) and 3 μg (lanes 4–7). M, molecular mass markers (from top to bottom): 120 kDa, 100 kDa, 85 kDa, 70 kDa, 60 kDa, 50 kDa and 40 kDa.
The ISOM activity of His6–ISOMp and the amidohydrolysing activity GAHp–His6 were virtually unaffected by UDP-GlcNAc, which is known to be a feedback inhibitor of eukaryotic GlcN-6-P synthase ([9] and references cited therein). In the presence of 2 mM UDP-GlcNAc, the Km of His6–ISOMp for Fru-6-P was 1.5±0.1 mM and the kcat was 0.53±0.04 min−1, whereas the Km of GAHp–His6 for GLUPA was 0.29±0.03 mM and the kcat was 0.48±0.05 min−1. On the other hand, UDP-GlcNAc lowered the GlcN-6-P-synthetic activity and amidohydrolysing activity of Gfa1p. For the former, the kcat value determined in the presence of 2 mM UDP-GlcNAc was 285±18 min−1, compared with 720±24 min−1 determined for the inhibitor-free enzyme. In the case of the amidohydrolysing activity, the effect was much stronger when Fru-6-P was also present in the incubation mixture. The kcat for GLUPA hydrolysis determined in the presence of 2 mM UDP-GlcNAc was 0.62±0.06 min−1 and in the presence of 2 mM UDP-GlcNAc and 10 mM Fru-6-P was 19.3±1.1 min−1, compared with 0.74±0.05 min−1 (−Fru-6-P) and 54.8±2.8 min−1 (+Fru-6-P) determined in the absence of UDP-GlcNAc. The Km values were not affected by UDP-GlcNAc.
Characterization of Δ218-283Gfa1p and the UDP-GlcNAc-binding site
The polypeptide chain of GlcN-6-P synthase of eukaryotic origin is generally 60–100 amino acids longer than its prokaryotic counterpart. Most of the inserts responsible for this elongation are located in the GAH region, and one of them is especially long in all known sequences [9]. In the C. albicans Gfa1p, the long insert comprises residues 218–283 (as shown in Figure 1). In order to find any evidence for the possible role of the 218–283 region of the enzyme, we constructed its truncated version, Δ218–283Gfa1p. The protein was isolated and characterized. The molecular mass of the native Δ218–283Gfa1p was found to be 290±2 kDa, whereas the molecular mass determined under denaturing condition was roughly four times lower (74±1 kDa), thus indicating that the removal of the 218–283 fragment does not affect the quaternary structure of Gfa1p.
The catalytic properties of the truncated enzyme were also characterized and compared with those of the wild-type enzyme. The measured activities are summarized in Table 3. It is evident that the truncated Gfa1p is not able to form GlcN-6-P but retains the amidohydrolysing and ISOM activities of the intact enzyme.
Table 3. Enzymatic activities of Gfa1p, its truncated and mutated versions.
The results are the means±S.D for three independent determinations.
| Amidohydrolysing activity (milliunits/mg) | ||||
|---|---|---|---|---|
| Enzyme version | −Fru-6-P | +Fru-6-P | ISOM activity (units/mg) | GlcN-6-P-synthetic activity (units/mg) |
| Gfa1p (wild-type) | 12±0.6 | 890±24 | 63.1±1.9 | 12±0.5 |
| Δ218–283Gfa1p | 11.6±0.2 | 12.5±0.7 | 61±2.2 | <0.01 |
| V711F | 13±0.5 | 940±18 | 62.6±1.3 | 0.021±0.003 |
| Δ709–712Gfa1p | 290±6 | 357±11 | 60.7±1.4 | <0.01 |
| W97F | 4.3±0.3 | 256±8 | 62.1±1.2 | 1.20±0.06 |
| W97G | 1.3±0.1 | 95±2 | 63.2±1.4 | 0.05±0.01 |
Gfa1p contains three tryptophan residues, at positions 97, 388 and 690. As the first one is located in the GAH domain and the remaining two are in the ISOM domain, we were able to use tryptophan fluorescence to study the interaction of GAHp–His6, His6–ISOMp, Δ218–283Gfa1p and Gfa1p with UDP-GlcNAc, by monitoring conformational changes induced in these proteins upon ligand binding. Emission spectra, taken at the tryptophan-specific excitation wavelength, were obtained and evaluated. We observed that the presence of UDP-GlcNAc strongly diminished the tryptophan-derived fluorescence of Gfa1p, thus indicating changes in the protein conformation, upon which at least one tryptophan residue becomes more exposed to the bulk solvent. The same effect was observed in the His6–ISOMp/UDP-GlcNAc spectrum, whereas the GAHp–His6 fluorescence was not affected by the ligand (results not shown). On the other hand, changes observed in the fluorescence spectrum of Δ218–283Gfa1p induced by UDP-GlcNAc (results not shown) were very similar to those found for the intact enzyme. The decrease in Gfa1p, Δ218–283Gfa1p and His6–ISOMp fluorescence induced by UDP-GlcNAc was ligand concentration-dependent and some subtle changes were observed even at the micromolar level (results not shown), corresponding to the level of the protein concentration, thus indicating a specific ligand binding.
One may therefore conclude that in the complete Gfa1p the conformational change triggered in the ISOM domain upon UDP-GlcNAc binding is subsequently transferred to the GAH domain through some interdomain interactions to affect there the amidohydrolysing activity, whereas the fragment containing residues 218–283 of the GAH domain does not seem to be involved in interactions with UDP-GlcNAc.
Interdomain communication and signalling
Structural studies on E. coli GlcN-6-P synthase revealed the presence of a 18 Å-long (1 Å=0.1 nm) hydrophobic channel (Figure 2) between the active sites of the GAH and ISOM domains, constituted by the residues Lys601, Ala602, Val605 and Val608 of the ISOM domain, and Arg26 and Trp74 of the GAH domain, with the additional participation of Lys503 and His504 of the neighbouring protein subunit in the functional dimer. The channel allows ammonia transfer from the GAH domain to the ISOM domain and is gated by the indole ring of Trp74 [5,8]. All the above-mentioned residues are strongly conserved in the known GlcN-6-P synthase sequences [9] and are also present in C. albicans Gfa1p as: Arg32, Trp97, Lys705, Ala706, Val709, Val711, Lys606 and His607. In the present study, we have constructed and characterized a few recombinant versions of Gfa1p, truncated or mutated at sites supposed to be crucial for the functioning of the putative channel. In particular, four C-terminal residues were removed to disturb the channel integrity, Val711 was replaced by phenylalanine to block the channel and finally Trp97 was exchanged for phenylalanine or glycine to reduce or remove a putative molecular gate, which is thought to be responsible for channel opening and closure. All the constructed mutants were tested for catalytic activity. The results are presented in Table 3. It is clear that the removal of residues 709–712 results in the complete elimination of the GlcN-6-P-synthetic activity of Gfa1p, whereas its ISOM activity remains unaffected. The ability to synthesize GlcN-6-P was not restored when we used ammonium chloride (10–50 mM) and 10 mM Fru-6-P as substrates (detailed results not shown). On the other hand, substitution of Trp97 for phenylalanine or glycine affected the amidohydrolysing activity and GlcN-6-P formation, but not the hexose phosphate isomerization. The effect was much stronger in the case of the W97G mutant, which lost almost all synthetic activity, and its ability to hydrolyse GLUPA was reduced to 10% of the original value. Interesting differences and changes in the amidohydrolysing activity, measured in the absence or presence of Fru-6-P, were noted. We observed a 70–80-fold increase of the amidohydrolysing activity of Gfa1p, Gfa1pV711F, Gfa1pW97F and Gfa1pW97G in the presence of Fru-6-P, whereas those of Δ709–712Gfa1p and Δ218–283Gfa1p were only slightly enhanced. These results indicate a possibility of an interdomain signalling in C. albicans Gfa1p. It should be noted that Mouilleron et al. [8] suggested that, in the E. coli GlcN-6-P synthase, interactions between residues constituting the C-terminal tail of ISOM, especially Thr606, and Cys1 and Asn98 of the GAH domain were responsible for a substantial enhancement of the amidohydrolase activity of GAH upon Fru-6-P binding at the ISOM domain. Our studies thus provided evidence supporting that suggestion, since the C-terminal tail–GAH interactions and, in consequence, the interdomain signalling, were apparently precluded upon deletion of residues 709–712. The negligible effect of Fru-6-P on amidohydrolysing activity of Δ218–283Gfa1p suggests that the interdomain signalling is also lost upon the deletion of residues 218–283. Rather unexpectedly, the amidohydrolysing activity of Δ709–712Gfa1p, measured in the absence of Fru-6-P, was nearly 25-fold higher than that of Gfa1p. This is surprising, since Δ709–712Gfa1p lacks a part of the C-terminal tail, including the Thr710 residue, corresponding to Thr606 of the bacterial GlcN-6-P synthase. Possibly some new interactions between the remaining part of the C-terminal tail and the GAH domain, which enhance the amidohydrolysing potential of the GAH domain, but which are practically independent of Fru-6-P binding, were created upon the removal of residues 709–712. Obviously, such an explanation is only speculative, unless the Gfa1p and Δ709–712Gfa1p structures are determined.
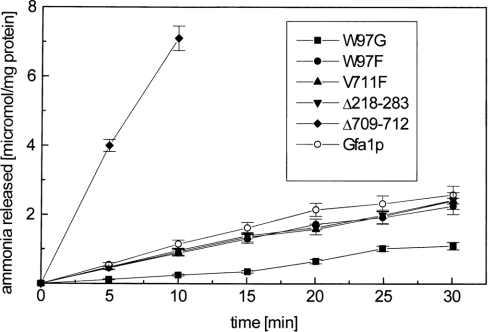
The results presented in Table 2 indicate that the Km values for the substrate in the amidohydrolysing activity of GAH–His6 and Gfa1p were not very different. This is in contrast with the previous observation by Isupov et al. [18], which was made using the bacterial GlcN-6-P synthase, where the Km for the whole enzyme was more than two orders of magnitude lower, as compared with that of the isolated domain. Teplyakov et al. [5] suggested that the higher affinity of the E. coli whole enzyme for glutamine was due to the interdomain interactions involving the Arg539 residue of the ISOM domain and the Q-loop (residues 73–80) of the GAH domain. Our results may thus indicate that such interactions affecting the glutamine binding do not exist in C. albicans Gfa1p, in spite of the fact that the above-mentioned residues are conserved in the fungal enzyme (as shown in Figure 1). The differences in the amidohydrolysing activity of the obtained muteins were of special interest. However, it must be kept in mind that the activity was measured with an artificial substrate. In order to check whether the observed phenomena take place also in the case of the physiological substrate, we measured ammonia release from L-glutamine in the absence of Fru-6-P for muteins and the wild-type enzyme. The results are shown in Figure 6. They confirm all the previous findings concerning the amidohydrolysing activity measured with GLUPA.
Figure 6. Time course of ammonia release from L-Gln, catalysed by Gfa1p and its mutants.
Enzyme samples were incubated with 10 mM L-Gln and the released ammonia was quantified with the Nessler's reagent. The results are shown as means±S.D. for experiments performed in triplicate.
The V711F mutant exhibited extremely low, but measurable, synthetic activity thus indicating that the introduced phenyl-ring effectively blocks the interdomain channel, and thus virtually prevents ammonia transfer from the GAH domain to the ISOM domain. The channel blockage did not influence the catalytic actions of either domain, since the measured respective activities were very much the same as those for the wild-type enzyme.
DISCUSSION
The results of the present study indicate that the two-domain structure, which has been demonstrated previously in the subunit of the bacterial GlcN-6-P synthase [5,18–20], is also a feature of the fungal enzyme and that the GAH domain functionality can be attributed to residues 1–345 of C. albicans Gfa1p, while the remaining part of the enzyme is catalytically active as the ISOM domain. The results of the present study also show that formation of the previously suggested homotetrameric structure of C. albicans GlcN-6-P synthase [14,21] is mediated by interactions between the ISOM domains, whereas the GAH domains do not seem to be involved in the oligomerization. Thus the quaternary structure of this enzyme is clearly different from that of its bacterial, homodimeric counterpart, as well as from that of the ‘sister’ class II amidotransferase, namely L-Gln:phosphoribosyl–pyrophosphate amidotransferase, for which it was previously found that the formation of the tetrameric structure was mediated by both domains [22].
Previous studies on fungal GlcN-6-P synthase provided kinetic evidence that the UDP-GlcNAc-binding site is different from the L-Gln- and D-Fru-6-P-binding sites [14,23]. The present results demonstrate that the inhibitor-binding site is located in the ISOM domain and that the binding of UDP-GlcNAc induces conformational changes in this domain. Such localization of the UDP-GlcNAc-binding site(s) confirms suggestions made by Chou [24] for the mammalian GlcN-6-P synthase, based on in silico homology modelling. The results of the present study also show that, despite the detected conformational changes, the UDP-GlcNAc binding to Gfa1p does not affect the ISOM activity of the ISOM domain, but lowers the amidohydrolysing activity of the GAH domain, and, as a consequence, the GlcN-6-P-synthetic activity of the entire enzyme. The structural basis for this effect will remain unclear until the exact location of the UDP-GlcNAc-binding site in the ISOM domain is identified.
The regulation of the GlcN-6-P synthase activity by enzymatic phosphorylation and dephosphorylation has been demonstrated previously for the mammalian and fungal enzymes [14,25–27]. The results of the present study prove that an increase of GlcN-6-P synthetic activity of the C. albicans Gfa1p, observed upon phosphorylation of the Ser208 residue of this enzyme, is most probably due to the enhancement of the amidohydrolysing activity of the GAH domain. The molecular mechanism of this effect will remain unknown until the structure of the fungal enzyme is solved. However, it seems worth mentioning that in the three-dimensional structure of the bacterial enzyme, the region corresponding to the PKA recognition site in the eukaryotic GlcN-6-P synthase is located close to the catalytic Cys1 residue. In particular, the hydroxyl group of Ser176 (homologous to Ser208 of the C. albicans enzyme) is only 5.4 Å from the protonated α-amino group of Cys1 [5,8,18]. If the same is true for the fungal enzyme, one may assume that the phosphorylated Ser208 may help keep Cys1 in the active conformation. However, the fungal GAH contains several inserts in the 1–208 region that are absent from the bacterial enzyme, which may also affect the relative positioning of Cys1 and Ser208.
A possible role for the residues 218–283 in C. albicans Gfa1p remains one of the particularly intriguing issues. Similar inserts, either smaller or slightly larger, are present in all known eukaryotic sequences, although they demonstrate very little, if any, sequence homology [9] that could suggest that the respective regions may not have any particular function. Our present results indicate that the 218–283 fragment is neither involved in UDP-GlcNAc binding nor in the formation of the quaternary structure. The fact that the Δ218–283Gfa1p lacks the GlcN-6-P-synthetic activity, can be, in our opinion, interpreted as a consequence of a possible improper relative orientation of the GAH and ISOM domains that prevents the assembly of the interdomain channel and thus precludes the ammonia transfer from the GAH domain to the ISOM domain. This suggestion seems to be additionally supported by the observed disturbance of the interdomain signalling.
The loss of the GlcN-6-P-synthetic activity of Gfa1p upon removal of the four C-terminal residues indicates that the integrity of the interdomain channel is crucial for the ammonia transfer from the GAH domain to the ISOM domain. Therefore it is very likely that the channel in Δ709–712Gfa1p is leaky and ammonia molecules, generated from L-Gln at the GAH domain active site, do not reach the ISOM domain active site, but are released to the bulk solvent. On the other hand, the escape route created by the 709–712 deletion seems to be unidirectional, since the truncated enzyme was still unable to use exogenous ammonia as the substrate for GlcN-6-P formation. However, one must keep in mind that GlcN-6-P synthase, unlike some other amidotransferases [6], does not possess any separate binding site for ammonia. Therefore the NH3 molecules from the bulk solvent, randomly interacting with enzyme molecules, may have little chance to enter the ISOM domain active site from a proper direction.
Finally, our present results provide evidence for the crucial role of the Trp97 residue, which is a counterpart of Trp74 of the E. coli enzyme. In our W97F and W97G mutants, this putative molecular gate was reduced in the former and virtually eliminated in the latter. These modifications have been shown to substantially decrease the GlcN-6-P-synthetic activity of the enzyme, by an order and more than two orders of magnitude respectively. In our opinion, this phenomenon may be well explained in light of the previous observations of Mouilleron et al. [8], who found that the indole ring of Trp74 in the E. coli enzyme, after rotation induced upon glutamine binding, opened the channel and at same time sealed its side-wall. We assume, therefore, that the channels in the W97F and W97G muteins of Gfa1p are leaky to some extent, resulting in lower efficiency of ammonia transfer from the GAH domain to the ISOM domain. Interestingly, both mutations resulted also in a slight decrease of the amidohydrolysing activity. This effect might be, in turn, caused by the fact that Trp74 in E. coli GlcN-6-P synthase, and possibly its counterparts in other enzyme variants, are parts of the Q-loop (residues 73–80 and 98–103 in the E. coli and C. albicans enzymes respectively), functioning as a lid that closes the entrance to the GAH domain active site when L-glutamine is bound [5,8]. The W97F and W97G mutations may distort the lid structure, thus making it less efficient as a ‘substrate keeper’.
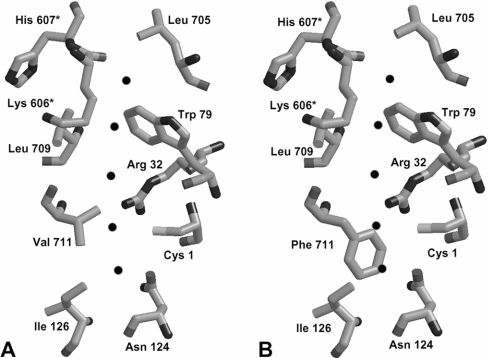
The V711F substitution appeared to create an additional molecular gate in the interdomain channel. This effect was in fact expected, as comparison of the structures of channel segments in V711F and the wild-type enzyme, made by molecular homology modelling (Figure 7), revealed that the phenyl ring should provide a steric hindrance to the channel. A similar phenomenon had been observed in the G359Y and G359F mutants of E. coli CPS (carbamoyl phosphate synthetase) [28,29], in the L109A mutant of E. coli CTP synthase [30] and in the L415A mutant of glutamine phosphoribosylpyrophosphate amidotransferase [31]. In all those cases, the partial reactions of glutamine hydrolysis and the final product formation became uncoupled, but both enzymes remained able to use exogenous ammonia as the substrate. It was postulated that the molecular channel in bacterial CPS becomes leaky upon Gly359 replacement with tyrosine or phenylalanine [29], whereas the channel in the CTP synthase was constricted upon the L109A exchange [30]. Our V711F mutant did not gain any ability to use ammonia for GlcN-6-P formation, but the partial activities were not affected. It seems therefore that in the V711F mutant, L-Gln or L-GLUPA is hydrolysed at the GAH domain, and the respective product, namely ammonia or p-nitroanilide, is released to the bulk solvent.
Figure 7. Blockage of the interdomain channel in Gfa1p upon V711F substitution.
(A) Computed structure created by homology modelling using E. coli GlcN-6-P synthase (1JXA) [5] as a template. (B) Putative result of V711F substitution. Residues marked with an asterisk (*) are from the neighbouring subunit. The channel is indicated by black spheres.
In conclusion, we were able to identify the separately expressed 1–345 and 346–712 fragments of C. albicans Gfa1p as the GAH domain and the ISOM domain respectively. The interactions between the ISOM domains are responsible for the formation of a homotetrameric structure of the native Gfa1p, whereas the GAH domain is not involved in protein oligomerization. The binding site for the feedback inhibitor UDP-GlcNAc is located in the ISOM domain. Binding of UDP-GlcNAc to the ISOM domain or phosphorylation of the Ser208 residue affects the amidohydrolysing activity of GAH and, in consequence, the GlcN-6-P synthetic activity of the whole enzyme. No specific function can be attributed to the 218–283 fragment of the GAH domain; however, its deletion strongly disrupts the interdomain communication and signalling. A similar effect is observed upon the removal of four C-terminal residues. The intramolecular channel connecting the GAH and ISOM domains is constituted by several hydrophobic residues and one of them, namely Trp97, functions as a molecular gate. The channel becomes leaky upon the deletion of amino acids 709–712, as well as upon the reduction of the molecular gate size. On the other hand, the channel can be blocked by V711F mutation.
Acknowledgments
We acknowledge the financial support of these studies from the Polish Ministry of Science and Higher Education (grant No P06A 036 25) and in part from the Faculty of Chemistry, Gdañsk University of Technology. We thank Dr. Marek Wojciechowski (Gdansk University of Technology, Poland) for his help with the molecular modelling.
References
- 1.Hebert L. F., Daniels M. C., Zhou J., Crook E. D., Turner R. L., Simmons S. T., Neidigh J. L., Zhu J. S., Baron A. D., McClain D.A. Overexpression of glutamine:fructose-6-phosphate amidotransferase in transgenic mice leads to insulin resistance. J. Clin. Invest. 1996;98:930–936. doi: 10.1172/JCI118876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borowski E. Novel approaches in the rational design of antifungal agents of low toxicity. Farmaco. 2000;55:206–208. doi: 10.1016/s0014-827x(00)00024-0. [DOI] [PubMed] [Google Scholar]
- 3.Badet B., Vermoote P., Haumont P. Y., Lederer F., LeGoffic F. Glucosamine synthetase from Escherichia coli: purification, properties and glutamine-utilizing site location. Biochemistry. 1987;26:1940–1948. doi: 10.1021/bi00381a023. [DOI] [PubMed] [Google Scholar]
- 4.Teplyakov A., Leriche C., Obmolova G., Badet B., Badet-Denisot M.-A. From Lobry de Bruyn to enzyme-catalysed ammonia channelling: molecular studies of D-glucosamine-6P synthase. Nat. Prod. Rep. 2002;19:60–69. doi: 10.1039/b103713g. [DOI] [PubMed] [Google Scholar]
- 5.Teplyakov A., Obmolova G., Badet B., Badet-Denisot M.-A. Channeling of ammonia in glucosamine-6-phosphate synthase. J. Mol. Biol. 2001;313:1093–1102. doi: 10.1006/jmbi.2001.5094. [DOI] [PubMed] [Google Scholar]
- 6.Massière F., Badet-Denisot M.-A. The mechanism of glutamine-dependent amidotransferases. Cell. Mol. Life Sci. 1998;54:205–222. doi: 10.1007/s000180050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leriche C., Badet-Denisot M.-A., Badet B. Affinity labeling of Escherichia coli glucosamine-6-phosphate synthase with a fructose 6-phosphate analog: evidence for proximity between the N-terminal cysteine and the fructose-6-phosphate-binding site. Eur. J. Biochem. 1997;245:418–422. doi: 10.1111/j.1432-1033.1997.00418.x. [DOI] [PubMed] [Google Scholar]
- 8.Mouilleron S., Badet-Denisot M.-A., Golinelli-Pimpaneau B. Glutamine binding opens the ammonia channel and activates glucosamine-6P synthase. J. Biol. Chem. 2006;281:4404–4412. doi: 10.1074/jbc.M511689200. [DOI] [PubMed] [Google Scholar]
- 9.Milewski S. Glucosamine-6-phosphate synthase – the multi-facets enzyme. Biochim. Biophys. Acta. 2002;1597:173–192. doi: 10.1016/s0167-4838(02)00318-7. [DOI] [PubMed] [Google Scholar]
- 10.Sachadyn P., Jedrzejczak R., Milewski S., Kur J., Borowski E. Purification to homogenity of Candida albicans glucosamine-6-phosphate synthase overexpressed in Escherichia coli. Prot. Expr. Purif. 2000;19:343–349. doi: 10.1006/prep.2000.1253. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J., Fritsch E. F., Maniatis T. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 12.Gabriel I., Olchowy J., Stanisławska-Sachadyn A., Mio T., Kur J., Milewski S. Phosphorylation of glucosamine-6-phosphate synthase is important but not essential for germination and mycelial growth of Candida albicans. FEMS Microbiol. Lett. 2004;235:73–80. doi: 10.1016/j.femsle.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Noltman E. A. Isolation of crystalline phosphoglucose isomerase from rabbit muscle. J. Biol. Chem. 1964;239:1545–1550. [PubMed] [Google Scholar]
- 14.Milewski S., Kuszczak D., Jedrzejczak R., Smith R. J., Brown A. J., Gooday G. W. Oligomeric structure and regulation of Candida albicans glucosamine-6- phosphate synthase. J. Biol. Chem. 1999;274:4000–4008. doi: 10.1074/jbc.274.7.4000. [DOI] [PubMed] [Google Scholar]
- 15.Olchowy J., Kur K., Sachadyn P., Milewski S. Construction, purification and functional characterisation of His-tagged Candida albicans glucosamine-6-phosphate synthase expressed in Escherichia coli. Prot. Expr. Purif. 2006;46:309–315. doi: 10.1016/j.pep.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 16.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Isupov M. N., Obmolova G., Butterworth S., Badet-Denisot M.-A., Badet B., Polikarpov I., Littlechild J. A., Teplyakov A. Substrate binding is required for assembly of the active conformation of the catalytic site in Ntn amidotransferases: evidence from the 1.8 Å structure of the glutaminase domain of glucosamine-6-phosphate synthase. Structure. 1996;4:801–810. doi: 10.1016/s0969-2126(96)00087-1. [DOI] [PubMed] [Google Scholar]
- 19.Denisot M.-A., LeGoffic F., Badet B. Glucosamine-6-phosphate synthase from Escherichia coli yields two proteins upon limited proteolysis: identification of the glutamine amidohydrolase and 2R ketose/aldose isomerase-bearing domains based on their biochemical properties. Arch. Biochem. Biophys. 1991;288:225–230. doi: 10.1016/0003-9861(91)90188-o. [DOI] [PubMed] [Google Scholar]
- 20.Teplyakov A., Obmolova G., Badet-Denisot M.-A., Badet B., Polikarpov I. Involvement of the C-terminus in intramolecular channeling in glucosamine-6-phosphate synthase: evidence from a 1.6 Å structure of the isomerase domain. Structure. 1998;6:1047–1055. doi: 10.1016/s0969-2126(98)00105-1. [DOI] [PubMed] [Google Scholar]
- 21.Olchowy J., Jedrzejczak R., Milewski S., Rypniewski W. Crystallization and preliminary X-ray analysis of the isomerase domain of glucosamine-6-phosphate synthase from Candida albicans. Acta Cryst. 2005;F61:994–996. doi: 10.1107/S174430910503318X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muchmore C. R. A., Krahn J. M., Kim J. H., Zalkin H., Smith J. L. Crystal structure of glutamine phosphoribisylpyrophosphate amidotransferase from Escherichia coli. Protein Sci. 1998;1:39–51. doi: 10.1002/pro.5560070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Endo A., Kakiki K., Misato T. Feedback inhibition of L-glutamine D-fructose 6-phosphate amidotransferase by uridine diphosphate N-acetylglucosamine in Neurospora crassa. J. Bacteriol. 1970;103:588–594. doi: 10.1128/jb.103.3.588-594.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou K.-C. Molecular therapeutic target for Type-2 diabetes. J. Proteome Res. 2004;3:1284–1288. doi: 10.1021/pr049849v. [DOI] [PubMed] [Google Scholar]
- 25.Chang Q., Su K., Baker J. R., Yang X., Paterson A. J., Kudlow J. E. Phosphorylation of human glutamine-fructose-6-phosphate amidotransferase by cAMP-dependent protein kinase at serine 205 blocks the enzyme activity. J. Biol. Chem. 2000;275:21981–21987. doi: 10.1074/jbc.M001049200. [DOI] [PubMed] [Google Scholar]
- 26.Hu Y., Riesland L., Paterson A. J., Kudlow J. E. Phosphorylation of mouse glutamine-fructose-6-phosphate amidotransferase 2 (GFAT2) by cAMP-dependent protein kinase increases the enzyme activity. J. Biol. Chem. 2004;279:29988–29993. doi: 10.1074/jbc.M401547200. [DOI] [PubMed] [Google Scholar]
- 27.Etchebehere L. C., Maia J. C. Phosphorylation-dependent regulation of amidotransferase during the development of Blastocladiella emersonii. Arch. Biochem. Biophys. 1989;272:301–310. doi: 10.1016/0003-9861(89)90223-3. [DOI] [PubMed] [Google Scholar]
- 28.Thoden J. B., Huang X., Raushel F. M., Holden H. M. Carbamoyl-phosphate synthetase: creation of an escape route for ammonia. J. Biol. Chem. 2002;277:39722–39727. doi: 10.1074/jbc.M206915200. [DOI] [PubMed] [Google Scholar]
- 29.Huang X., Raushel F. M. An engineered blockage within the ammonia tunnel of carbamoyl phosphate synthase prevents the use of glutamine as a substrate but not ammonia. Biochemistry. 2000;39:3240–3247. doi: 10.1021/bi9926173. [DOI] [PubMed] [Google Scholar]
- 30.Lunn F. A., Bearne S. L. Alternative substrates for wild-type and L109A E. coli CTP synthases: kinetic evidence for a constricted ammonia tunnel. Eur. J. Biochem. 2004;271:4204–4212. doi: 10.1111/j.1432-1033.2004.04360.x. [DOI] [PubMed] [Google Scholar]
- 31.Bera A. K., Smith J. L., Zalkin H. Dual role for the glutamine phosphoribosylpyrophosphate amidotransferase ammonia channel: interdomain signaling and intermediate channeling. J. Biol. Chem. 2000;275:7975–7979. doi: 10.1074/jbc.275.11.7975. [DOI] [PubMed] [Google Scholar]