Figure 3.
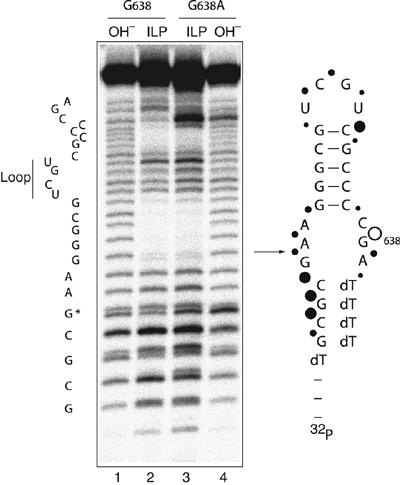
Comparison of the conformations of the natural and G638A substrates by in-line probing. Versions of natural sequence and G638A substrates with 5′ and 3′ terminal extensions of deoxyribonucleotides were synthesized to improve the electrophoretic resolution of the RNA sections. Radioactively 5′-32P-labelled substrates were incubated in standard VS buffer at 25°C for 40 h. Cleavages were analysed by electrophoresis in a 20% polyacrylamide gel under denaturing conditions. Tracks 1 and 4, base cleavage of natural and G638A substrates, respectively and tracks 2 and 3, in-line probing analysis of natural and G638A substrates, respectively. The scheme shows the sequence of the natural substrate, with the arrow indicating the position of ribozyme cleavage. The positions of sensitivity to in-line probing are indicated by filled circles, the size of which reflects the extent of cleavage. The open circle shows the position of the phosphodiester linkage that is more sensitive in the G638A substrate; note that this position also exhibits enhanced base cleavage. The shorter fragments migrate as doublets, due to resolution of the cyclic 2′3′-phosphates and their products of hydrolysis during the long incubation.
