Figure 6.
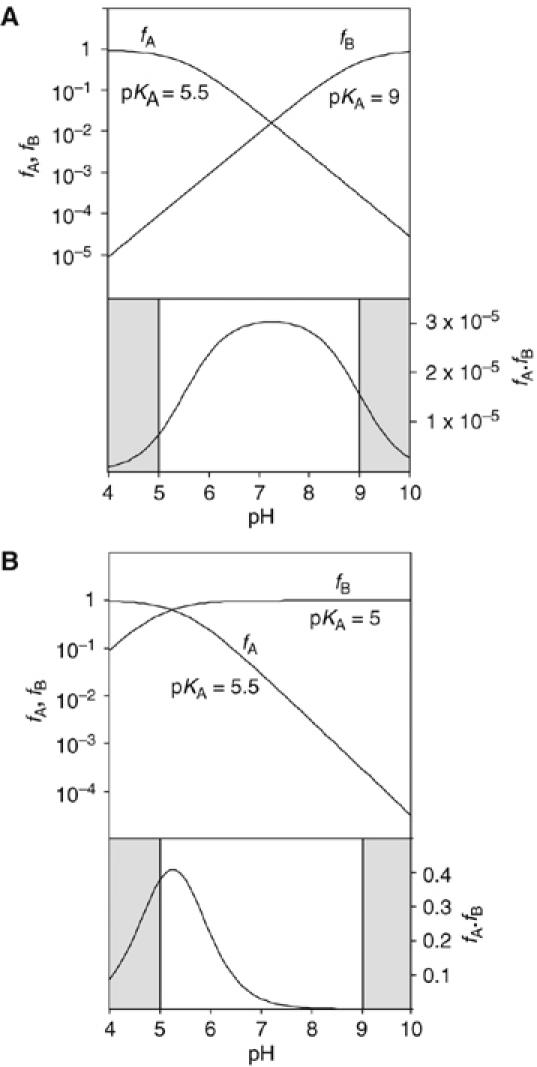
Calculated pH dependence of the cleavage reaction of the VS ribozyme as a function of base pKA, assuming general acid–base catalysis. The fractions of protonated acid fA, unprotonated base fB and their product fA·fB have been calculated and plotted as a function of pH, following the approach of Bevilacqua (2003). The shaded sections are the regions of pH not accessible to experimental study. Reaction rate should be proportional to the fraction of ribozyme in the appropriate state of protonation, that is, fA·fB. Note that in these graphs, fA and fB are plotted on a log10 scale (left), whereas fA·fB is plotted on a linear scale (right). (A) Plot for pKA values of 5.5 and 9 for the acid and base, respectively. This might correspond to the natural ribozyme, assuming that the acid is an adenine with an elevated pKA and the base is a guanine with a slightly reduced pKA. The predicted reaction rate profile is a broad bell shape, with a maximum close to neutrality. (B) Plot for pKA values of 5.5 and 5 for the acid and base, respectively. This situation could emerge if the guanine were replaced by a nucleobase of significantly lower pKA (e.g. adenine or DAP). The reaction rate is predicted to exhibit a marked increase at low pH, with a maximum that is just detectable for these values.
