Figure 1.
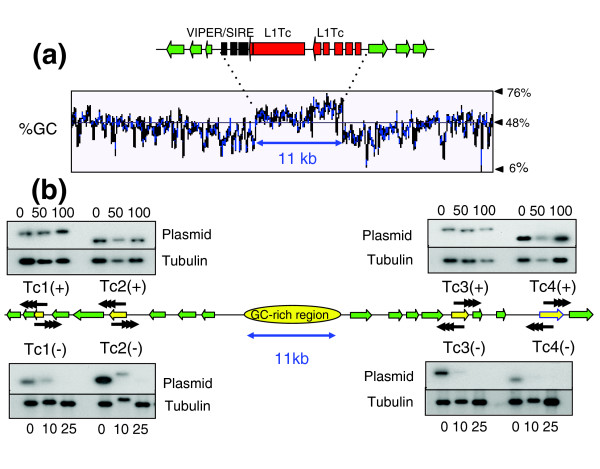
Functional mapping of the putative centromere on T. cruzi chromosome 1. (a) Organization of the GC-rich strand-switch region. Green arrows identify ORFs in the polycistronic gene clusters and the implied direction of transcription. The degenerate retrotransposon-like VIPER/SIRE element (black) and L1Tc autonomous retroelements (red) are indicated. The %GC content was determined by the Artemis 7 program [38]. (b) Mitotic stability of truncated chromosomes. Sequences used for fragmentation (Tc1-Tc4) are indicated by yellow arrows. Vectors were targeted in both directions (+/-), with black arrowheads representing the positions and orientations of de novo telomeres after fragmentation (see Additional data files 1-3 for further details). Clones with truncated chromosomes were grown in the absence of G418 for the generations indicated above or below the corresponding track. Genomic DNA was ScaI digested, Southern blotted, probed with plasmid DNA, then re-hybridized with β-tubulin as a loading control.
