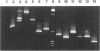
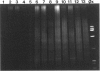
Abstract
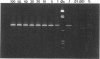
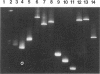
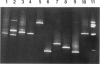
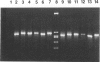
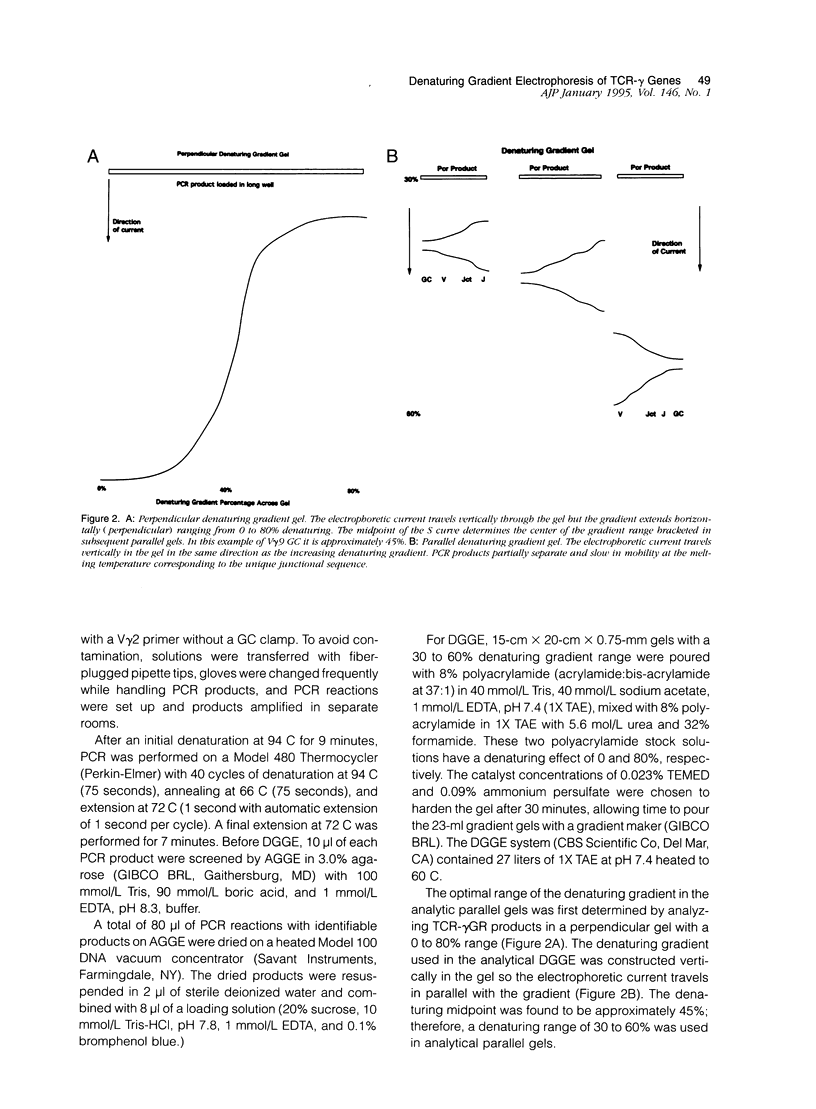
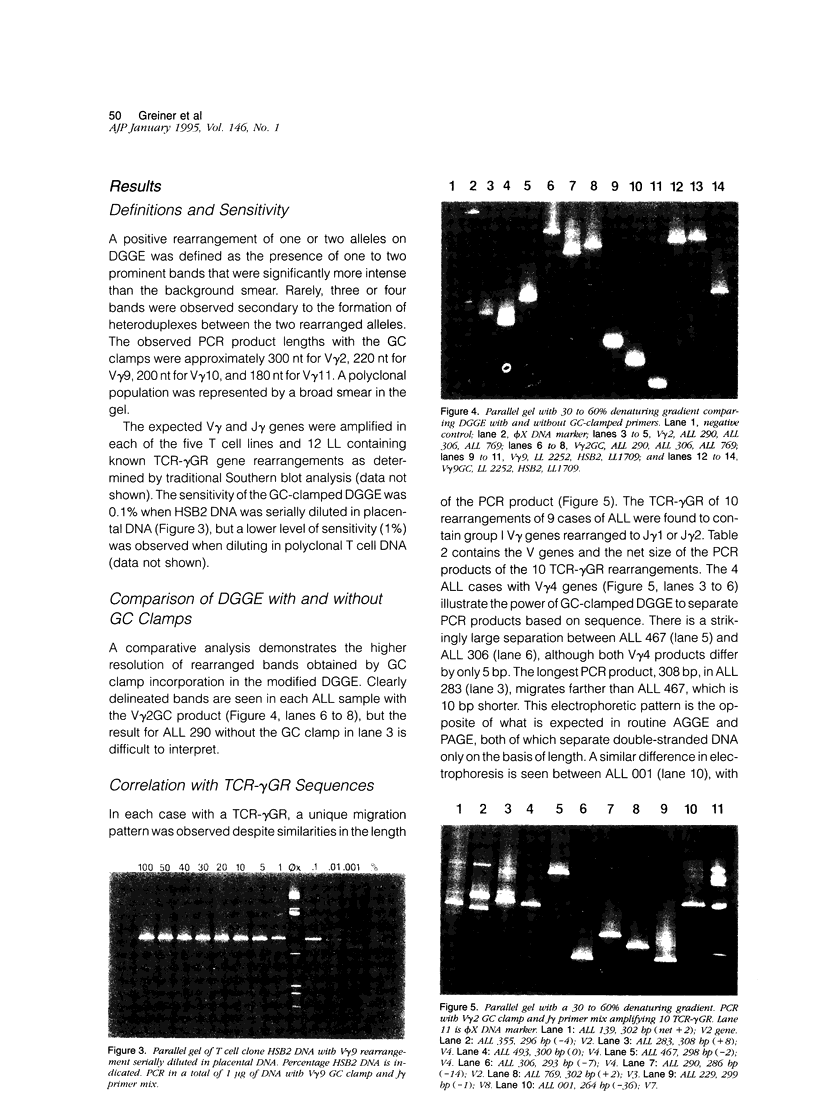
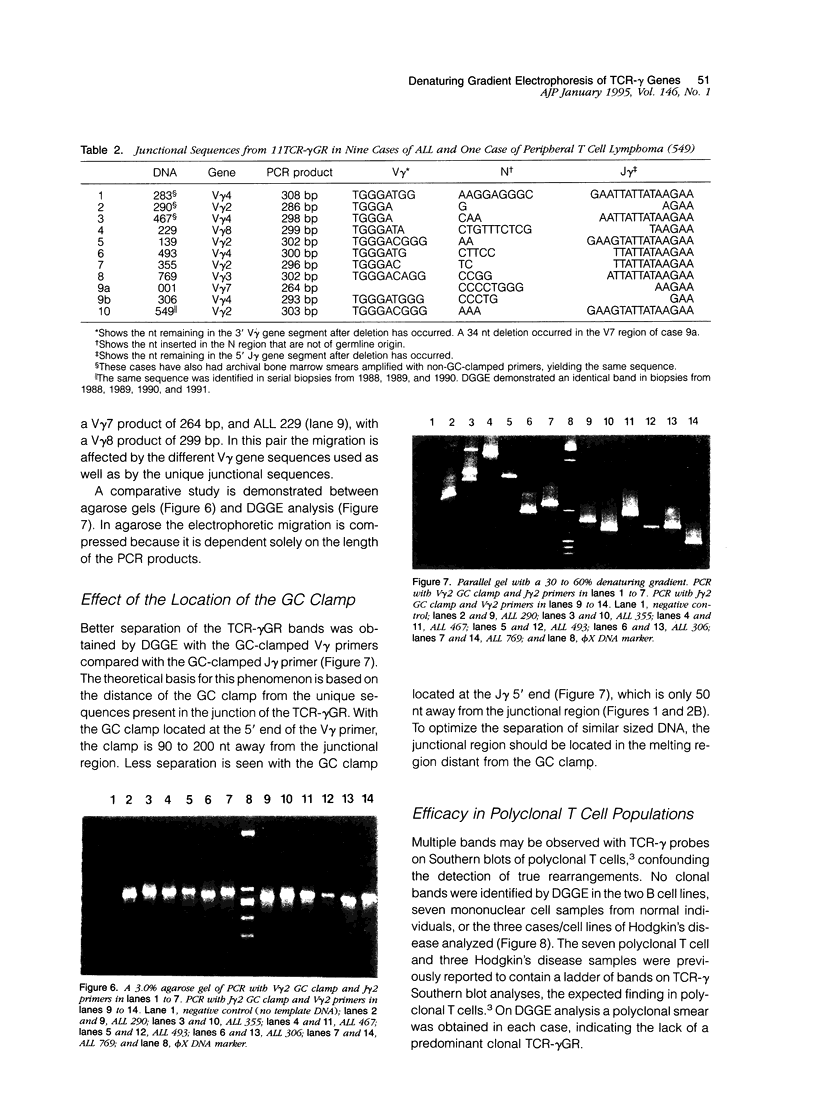
We describe a modified denaturing gradient gel electrophoresis (DGGE) procedure with a 40-nucleotide GC clamp in the polymerase chain reaction to improve resolution in amplifying T cell receptor-gamma (TCR-gamma) rearrangements. DNA from 46 cases of lymphoblastic leukemia/lymphoma, 5T cell lines, 2 B cell lines, 7 normal lymphocytes, and 3 cases of Hodgkin's disease was amplified by polymerase chain reaction. In addition, 20 cases of paraffin-embedded T cell lymphomas and 5 cases of reactive hyperplasia were also studied. Clonal TCR-gamma rearrangements were identified on DGGE by the presence of a predominant band. Results obtained from 5 T cell lines and 12 lymphoblastic leukemia/lymphomas containing known TCR-gamma gene rearrangements revealed 100% concordance in detecting clonal rearrangements between DGGE and traditional Southern blot analysis. Of the remaining 34 lymphoblastic leukemia/lymphoma cases studied by DGGE alone, 30 were positive. DGGE analysis of 10 lymphoblastic leukemia/lymphoma cases with known group IV gamma to J gamma 1 or J gamma 2 rearrangement sequences confirmed that the electrophoretic migration was dependent on the tumor-specific rearranged TCR-gamma sequence. In addition, 17 of 20 cases of paraffin-embedded T cell lymphomas were positive by DGGE, 6 of which had the clonal population also identified in fresh tissue DNA. DGGE analysis of GC-clamped polymerase chain reaction products can provide a way to more accurately detect TCR-gamma clonality of lymphoid tumors and can be applied to archival tissues.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrams E. S., Murdaugh S. E., Lerman L. S. Comprehensive detection of single base changes in human genomic DNA using denaturing gradient gel electrophoresis and a GC clamp. Genomics. 1990 Aug;7(4):463–475. doi: 10.1016/0888-7543(90)90188-z. [DOI] [PubMed] [Google Scholar]
- Bahler D. W., Berry G., Oksenberg J., Warnke R. A., Levy R. Diversity of T-cell antigen receptor variable genes used by mycosis fungoides cells. Am J Pathol. 1992 Jan;140(1):1–8. [PMC free article] [PubMed] [Google Scholar]
- Bourguin A., Tung R., Galili N., Sklar J. Rapid, nonradioactive detection of clonal T-cell receptor gene rearrangements in lymphoid neoplasms. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8536–8540. doi: 10.1073/pnas.87.21.8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A., Huck S., Ghanem N., Lefranc M. P., Rabbitts T. H. New subgroups in the human T cell rearranging V gamma gene locus. EMBO J. 1987 Jul;6(7):1945–1950. doi: 10.1002/j.1460-2075.1987.tb02456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudie R. B., Karim S. N., Mills K., Alcorn M., Lee F. D. A sensitive method of screening for dominant T cell clones by amplification of T cell gamma gene rearrangements with the polymerase chain reaction. J Pathol. 1990 Nov;162(3):191–196. doi: 10.1002/path.1711620304. [DOI] [PubMed] [Google Scholar]
- Hansen-Hagge T. E., Yokota S., Bartram C. R. Detection of minimal residual disease in acute lymphoblastic leukemia by in vitro amplification of rearranged T-cell receptor delta chain sequences. Blood. 1989 Oct;74(5):1762–1767. [PubMed] [Google Scholar]
- Heller M. J., Burgart L. J., TenEyck C. J., Anderson M. E., Greiner T. C., Robinson R. A. An efficient method for the extraction of DNA from formalin-fixed, paraffin-embedded tissue by sonication. Biotechniques. 1991 Sep;11(3):372-4, 376-7. [PubMed] [Google Scholar]
- Huck S., Lefranc M. P. Rearrangements to the JP1, JP and JP2 segments in the human T-cell rearranging gamma gene (TRG gamma) locus. FEBS Lett. 1987 Nov 30;224(2):291–296. doi: 10.1016/0014-5793(87)80472-6. [DOI] [PubMed] [Google Scholar]
- Imrie F., Karim S. N., Goudie R. B. Error in reported frequency of dominant T-cell receptor V gamma 8 gene rearrangements in T-cell lymphomas. J Pathol. 1992 Apr;166(4):417–418. doi: 10.1002/path.1711660416. [DOI] [PubMed] [Google Scholar]
- Knowles D. M. Immunophenotypic and antigen receptor gene rearrangement analysis in T cell neoplasia. Am J Pathol. 1989 Apr;134(4):761–785. [PMC free article] [PubMed] [Google Scholar]
- LeFranc M. P., Forster A., Baer R., Stinson M. A., Rabbitts T. H. Diversity and rearrangement of the human T cell rearranging gamma genes: nine germ-line variable genes belonging to two subgroups. Cell. 1986 Apr 25;45(2):237–246. doi: 10.1016/0092-8674(86)90388-0. [DOI] [PubMed] [Google Scholar]
- Lefranc M. P., Forster A., Rabbitts T. H. Rearrangement of two distinct T-cell gamma-chain variable-region genes in human DNA. 1986 Jan 30-Feb 5Nature. 319(6052):420–422. doi: 10.1038/319420a0. [DOI] [PubMed] [Google Scholar]
- Lorenzen J., Jux G., Zhao-Höhn M., Klöckner A., Fischer R., Hansmann M. L. Detection of T-cell clonality in paraffin-embedded tissues. Diagn Mol Pathol. 1994 Jun;3(2):93–99. doi: 10.1097/00019606-199406000-00005. [DOI] [PubMed] [Google Scholar]
- Lutz C. T., Galles M. E., Kemp J. D., Goeken J. A., Dick F. R. Kappa immunoglobulin light chain gene rearrangement in a T-lineage chronic lymphocytic leukemia. Am J Clin Pathol. 1990 May;93(5):702–705. doi: 10.1093/ajcp/93.5.702. [DOI] [PubMed] [Google Scholar]
- Macintyre E. A., d'Auriol L., Duparc N., Leverger G., Galibert F., Sigaux F. Use of oligonucleotide probes directed against T cell antigen receptor gamma delta variable-(diversity)-joining junctional sequences as a general method for detecting minimal residual disease in acute lymphoblastic leukemias. J Clin Invest. 1990 Dec;86(6):2125–2135. doi: 10.1172/JCI114951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K. P., Sloane J. P., Kabarowski J. H., Matutes E., Wiedemann L. M. A simplified method of detection of clonal rearrangements of the T-cell receptor-gamma chain gene. Diagn Mol Pathol. 1992 Sep;1(3):173–179. [PubMed] [Google Scholar]
- McCarthy K. P., Sloane J. P., Kabarowski J. H., Matutes E., Wiedemann L. M. The rapid detection of clonal T-cell proliferations in patients with lymphoid disorders. Am J Pathol. 1991 Apr;138(4):821–828. [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Maniatis T., Lerman L. S. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- Neale G. A., Menarguez J., Kitchingman G. R., Fitzgerald T. J., Koehler M., Mirro J., Jr, Goorha R. M. Detection of minimal residual disease in T-cell acute lymphoblastic leukemia using polymerase chain reaction predicts impending relapse. Blood. 1991 Aug 1;78(3):739–747. [PubMed] [Google Scholar]
- Orita M., Suzuki Y., Sekiya T., Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989 Nov;5(4):874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sheffield V. C., Cox D. R., Lerman L. S., Myers R. M. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci U S A. 1989 Jan;86(1):232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield V. C., Fishman G. A., Beck J. S., Kimura A. E., Stone E. M. Identification of novel rhodopsin mutations associated with retinitis pigmentosa by GC-clamped denaturing gradient gel electrophoresis. Am J Hum Genet. 1991 Oct;49(4):699–706. [PMC free article] [PubMed] [Google Scholar]
- Slack D. N., McCarthy K. P., Wiedemann L. M., Sloane J. P. Evaluation of sensitivity, specificity, and reproducibility of an optimized method for detecting clonal rearrangements of immunoglobulin and T-cell receptor genes in formalin-fixed, paraffin-embedded sections. Diagn Mol Pathol. 1993 Dec;2(4):223–232. [PubMed] [Google Scholar]
- Tamura N., Holroyd K. J., Banks T., Kirby M., Okayama H., Crystal R. G. Diversity in junctional sequences associated with the common human V gamma 9 and V delta 2 gene segments in normal blood and lung compared with the limited diversity in a granulomatous disease. J Exp Med. 1990 Jul 1;172(1):169–181. doi: 10.1084/jem.172.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. J., Rowe D., Williamson I. K., Christmas S. E., Proctor S. J., Middleton P. G. Detection of T-cell receptor gamma chain V gene rearrangements using the polymerase chain reaction: application to the study of clonal disease cells in acute lymphoblastic leukemia. Blood. 1991 May 1;77(9):1989–1995. [PubMed] [Google Scholar]
- Trainor K. J., Brisco M. J., Wan J. H., Neoh S., Grist S., Morley A. A. Gene rearrangement in B- and T-lymphoproliferative disease detected by the polymerase chain reaction. Blood. 1991 Jul 1;78(1):192–196. [PubMed] [Google Scholar]
- Uppenkamp M., Andrade R., Sundeen J., Raffeld M., Coupland R., Cossman J. Diagnostic interpretation of T gamma gene rearrangement: effect of polyclonal T cells. Hematol Pathol. 1988;2(1):15–24. [PubMed] [Google Scholar]
- Uppenkamp M., Pittaluga S., Lipford E. H., Cossman J. Limited diversity and selection of rearranged gamma genes in polyclonal T cells. J Immunol. 1987 Mar 1;138(5):1618–1620. [PubMed] [Google Scholar]
- Volkenandt M., Soyer H. P., Cerroni L., Koch O. M., Atzpodien J., Kerl H. Molecular detection of clone-specific DNA in hypopigmented lesions of a patient with early evolving mycosis fungoides. Br J Dermatol. 1993 Apr;128(4):423–428. doi: 10.1111/j.1365-2133.1993.tb00203.x. [DOI] [PubMed] [Google Scholar]
- Volkenandt M., Soyer H. P., Kerl H., Bertino J. R. Development of a highly specific and sensitive molecular probe for detection of cutaneous lymphoma. J Invest Dermatol. 1991 Jul;97(1):137–140. doi: 10.1111/1523-1747.ep12479308. [DOI] [PubMed] [Google Scholar]
- Volkenandt M., Wienecke R., Koch O. M., Buer J., Probst M., Atzpodien J., Horikoshi T., Danenberg K., Danenberg P., Bertino J. R. Conformational polymorphisms of cRNA of T-cell-receptor genes as a clone-specific molecular marker for cutaneous lymphoma. J Invest Dermatol. 1993 Oct;101(4):514–516. doi: 10.1111/1523-1747.ep12365889. [DOI] [PubMed] [Google Scholar]
- d'Auriol L., Macintyre E., Galibert F., Sigaux F. In vitro amplification of T cell gamma gene rearrangements: a new tool for the assessment of minimal residual disease in acute lymphoblastic leukemias. Leukemia. 1989 Feb;3(2):155–158. [PubMed] [Google Scholar]