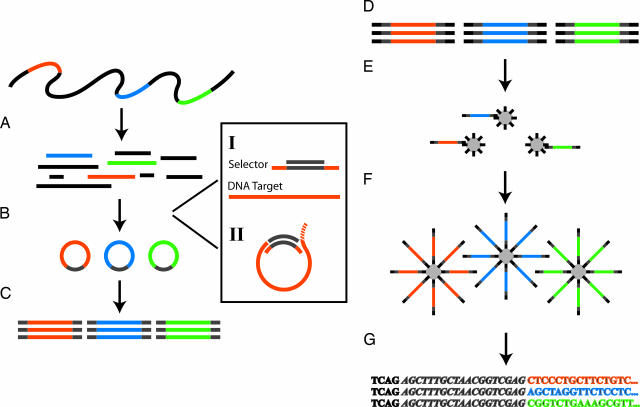
Fig. 1.
The selector and sequencing assay. (A) A DNA sample is digested to defined fragments by using restriction enzyme(s). The color bars represent the targets of interest. (B) Targeted circularization is performed by using selectors. (I) Selectors contain two oligonucleotides: a selector probe and a general vector oligonucleotide. The selector probe has two single-stranded target-complementary end sequences (orange) that are linked by a general sequence motif (gray) and the vector oligonucleotide that is complementary to the general sequence motif in the selector probes (gray). (II) The circularization reaction can be carried out by using two different approaches. Either both ends of the selected fragment connect to the vector oligonucleotide by hybridizing and ligation or the vector oligonucleotide forms a branched structure in an optional position at the 5′ end of the fragment. This latter structure is recognized and processed by the added endonucleolytic enzyme, forming ends suitable for ligation. (C) The circles are amplified in a multiplex PCR by using a primer pair complementary to the general vector sequence introduced in every circle. (D) The first step in the 454-sequencing procedure is to attach general, 454-optimized, adaptor sequences to each end of each PCR product. (E) The PCR products are separated into single strands and bound to beads in limiting dilutions, resulting in one unique fragment per bead. (F) The beads are clonally amplified in droplets of an oil-emulsion-based PCR, resulting in beads carrying millions of target sequences. (G) The beads are finally deposited into picoliter-sized wells, one bead per well, where solid-phase pyrosequencing is performed and monitored.