Abstract
Multiphoton excitation fluorescent microscopy is a laser-based technology that allows subcellular resolution of native tissues in situ. We have recently applied this technology to the structural and photochemical imaging of cultured glioma cells and experimental gliomas ex vivo. We demonstrated that high microanatomical definition of the tumor, invasion zone, and normal adjacent brain can be obtained down to single-cell resolution in unprocessed tissue blocks. In this study, we used multiphoton excitation and four-dimensional microscopy to generate fluorescence lifetime maps of the murine brain anatomy, experimental glioma tissue, and biopsy specimens of human glial tumors. In murine brain, cellular and non-cellular elements of the normal anatomy were identified. Distinct excitation profiles and lifetimes of endogenous fluorophores were identified for specific brain regions. Intracranial grafts of human glioma cell lines in mouse brain were used to study the excitation profiles and fluorescence lifetimes of tumor cells and adjacent host brain. These studies demonstrated that normal brain and tumor could be distinguished on the basis of fluorescence intensity and fluorescence lifetime profiles. Human brain specimens and brain tumor biopsies were also analyzed by multiphoton microscopy, which demonstrated distinct excitation and lifetime profiles in glioma specimens and tumor-adjacent brain. This study demonstrates that multiphoton excitation of autofluorescence can distinguish tumor tissue and normal brain based on the intensity and lifetime of fluorescence. Further technical developments in this technology may provide a means for in situ tissue analysis, which might be used to detect residual tumor at the resection edge.
Keywords: glioma, glioma invasion, fluorescence lifetime imaging, four-dimensional microscopy, multiphoton excitation fluorescence microscopy
Multiphoton microscopy uses near-infrared femtosecond laser pulses to excite endogenous intra- and extracellular fluorophores in a femtoliter target volume (König, 2000). The fluorescence of the excited endogenous fluorophores can be detected by a photomultiplier and may be reconstructed into three-dimensional intensity images of native target tissues at a subcellular resolution without the need for contrast-enhancing markers. In a conceptual study using experimental gliomas, we have recently demonstrated high anatomical definition of the tumor parenchyma, the invasion zone, and normal adjacent brain in unprocessed tissue blocks by multiphoton excitation autofluorescence microscopy (Leppert et al., 2006). Morphological characteristics of individual cell types could be identified at a single-cell level down to resolution of cellular organelles. This technology, however, is not limited to anatomical and structural imaging. Picosecond time-resolved detection of the photons emitted from multiphoton-excited fluorophores may be used to analyze the lifetime of the autofluorescence, which is the average time between excitation and emission of the fluorescence (Becker et al., 2001; Xu et al., 1996a, 1996b). Using specific excitation wavelengths, fluorescence lifetime imaging (four-dimensional microscopy) can selectively excite and detect endogenous molecular fluorophores by their excitation spectra and their fluorescence lifetime. Such biochemical imaging by multiphoton microscopy has been shown to distinguish extracellular matrix components such as elastic fibers from collagen in human skin (König et al., 2005) and has facilitated selective excitation of melanin (Teuchner et al., 1999). Recently, our analysis of the relationship between the laser excitation wavelength and the lifetime of excitable endogenous fluorophores in cells derived from tumors of different histotypes has suggested individual fluorescence lifetime profiles for distinct cell types. We have further shown that time-resolved measurements of fluorescence lifetimes distinguish tumor cells from normal brain parenchyma (Leppert et al., 2006).
In the present study, we used multiphoton excitation to generate color-coded fluorescence lifetime images of the murine brain anatomy, experimental glioma tissue, and biopsy specimens of human glial tumors. In murine brain, cellular and noncellular elements of the normal brain anatomy were identified, which showed distinct excitation profiles of endogenous fluorophores and a distinct spectrum of fluorescence lifetimes. We used intracranial grafts of human glioma cell lines in mouse brain to study the excitation profiles and fluorescence lifetimes of tumor cells and the adjacent host brain. These studies demonstrated that normal brain and tumor could be distinguished based on fluorescence intensity and distinct excitation/lifetime profiles. Unprocessed tissue blocks of human brain specimens and brain tumor biopsy specimens analyzed by multiphoton excitation also demonstrated distinct excitation/lifetime profiles in glioma specimens compared with normal brain.
Materials and Methods
Multiphoton Imaging System
For multiphoton excitation of endogenous fluorophores in experimental gliomas, we used the DermaInspect in vivo imaging system (JenLab, Jena, Germany). The system contains a solid-state, mode-locked 80-MHz titanium:sapphire laser (MaiTai, Spectra Physics, Darmstadt, Germany) with a tuning range of 710–920 nm, a mean laser output of >900 mW at 800 nm, and a 75-fs pulse width. The scanning module contains a motorized beam attenuator, a shutter, and a two-axis galvoscanner. A piezo-driven 40× focusing optic with a 1.3 numerical aperture and 140-μm working distance (Plan Neofluar, Zeiss, Göttingen, Germany) was used to study native brain and tumor tissue. The autofluorescence signal was detected by a photomultiplier tube module (H7732-01, Hamamatsu, Herrsching, Germany) after passing a beam splitter and a short-pass filter (BG39, Schott, Mainz, Germany).
Time-Resolved Autofluorescence Measurements
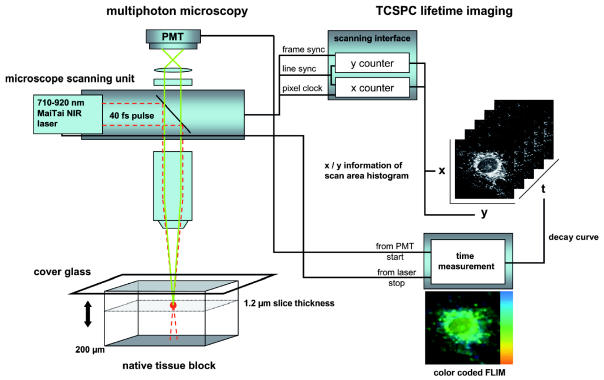
Fluorescence lifetime images were measured by time-correlated single-photon counting (Fig. 1). A photomultiplier module (PMH-100-0, Becker & Hickl, Berlin, Germany) detected the fluorescence photons emitted by the tissue. The start signal for the photomultiplier and stop signals provided by the laser were processed by a PC-based single-photon counting board (SPC 830, Becker & Hickl), which allowed count rates of up to 8 × 106 photons/s. The single-photon counting board was synchronized with the spatial beam position, which was calculated from signals of the galvoscanner. Spatially resolved autofluorescence decay curves were recorded for 256 × 256 pixels per image field, which typically was 150 μm. The depth of the excitation volume typically was less than 1 μm. Curve fitting of a single exponential decay curve, including a deconvolution with the time response of the system (SPCImage 2.6, Becker & Hickl), was used to calculate a mean fluorescence lifetime for each pixel, which was displayed in color-coded images (Becker et al., 2001). The accuracy of the measurements can be judged by the scattering of the measured values. Under optimal conditions when only the shot noise of the photons determines the relative error of the measured lifetimes, it is approximated by 2 divided by the square root of the number of detected photons, which was between 100 and a few thousand per pixel during the measurements (Köllner and Wolfrum, 1992). Therefore, errors of up to 10% are expected. To analyze the fluorescence lifetimes of endogenous fluorophores within specific cellular compartments, regions of interest were defined and the analysis was performed in at least three regions of similar compartments. The fluorescence lifetime for each region of interest was determined. The data are reported as the means of triplicate determinations, and the fluorescence lifetimes are plotted as a function of the excitation wavelength.
Fig. 1.
Schematic presentation of multiphoton microscopy of native central nervous system tissue and glioma tissue.
Orthotopic Glioma Mouse Model, Tumor Specimens, and Histology
The human glioblastoma–derived cell lines G-28, G-112, and U87 were grown in minimum essential medium containing 10% fetal calf serum. For intracranial implantation in nude NMRI mice, cells were expanded and harvested in log-phase growth by trypsinization. Cells were washed in PBS three times and then resuspended at a concentration of 2 × 104/μl. All procedures were performed in accordance with regulations of the Animal Care and Use Committee of the University Hospital of Schleswig-Holstein (permit 30/o/03). Mice were anesthetized by peritoneal injection of ketamine/xylazine solution (200 mg ketamine and 20 mg xylazine in 17 ml saline) at 0.15 mg/10 g body weight; the cranium was fixed in a stereotactic frame (TSE Systems, Bad Homburg, Germany). A 1-mm bur hole was drilled 3 mm lateral to the bregma, and a stereotactic implantation of 3 μl of cell suspension injected over 3 min was placed in an area corresponding to the internal capsule 0.5 mm below the fiber tracts of the corpus callosum. After implantation, 50 mg/kg novaminsulfone was administered subcutaneously, and 1 mg/ml novaminsulfone was added to the drinking water for three days. Four weeks after implantation, tumor-bearing brains were explanted following a lethal intraperitoneal injection of 50 mg/kg xylazine and 350 mg/kg ketamine. The specimens were processed on ice, and the brains were divided into two tissue blocks at a coronal plane using a scalpel. The tissue samples were placed in a humidified biopsy chamber (MiniCeM, JenLab) adherent to a 0.17-μm cover glass and imaged. Following the imaging studies, the specimens were fixed in formalin, and the tissue blocks were sectioned parallel to the optical imaging plane and embedded in paraffin; 5-μm sections were cut and stained with hematoxylin and eosin.
Results
Time-Resolved Autofluorescence Imaging of Murine Brain by Multiphoton Microscopy
Recently, we have shown that glioma cells in culture and cells derived from different histotypes differ in their multiphoton excitation/fluorescence lifetime profile, suggesting that multiphoton microscopy and fluorescence lifetime imaging may provide some cell-type specificity and a means of identifying glioma cells in brain tissue (Leppert et al., 2006). We therefore imaged the normal anatomy of native mouse brain tissue blocks and analyzed the excitability of fluorescence lifetime as a function of the excitation wavelength.
From specific areas of interest, lifetime images were obtained using increasing excitation wavelengths from 720 to 780 nm at increments of 10 nm. The distribution of fluorescence lifetime was color coded using a continuous spectrum of red (short lived) to blue (long lived). The lifetime of homogeneous areas, cells, or organelles such as the nucleus or highly autofluorescent granula were analyzed separately in some specimens. On the basis of these parameters, graphs were plotted displaying the lifetime of specific regions of interest as a function of the excitation wavelength (Fig. 2).
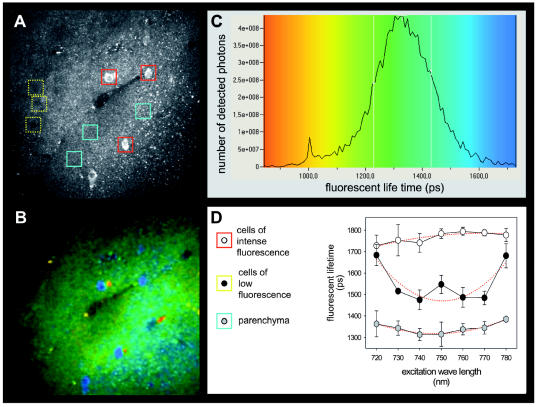
Fig. 2.
Multiphoton microscopy of normal mouse basal ganglia. (A) Intensity image of the autofluorescence signal, demonstrating cells of intense fluorescence with low-intensity nuclei. Near these intensely fluorescing cells, nuclei of cells with low-signal-intensity cytoplasm could be identified. Acellular areas of the parenchyma were selected based on three-dimensional stacks of images demonstrating no cellular nuclei above or below the plane of analysis. (B) Corresponding color-coded fluorescence lifetime image generated at an excitation wavelength of 750 nm. (C) Color-coded distribution of fluorescence lifetimes at 750 nm within the whole image frame, shown in picoseconds. (D) Excitation/lifetime profile of different areas of interest framed in A. This analysis demonstrated that high-intensity fluorescence cells not only showed a longer fluorescence lifetime than did low-intensity cells but also showed distinct excitation/lifetime profiles.
Normal mouse brain contained several anatomical and microanatomical structures readily identified by multiphoton microscopy. Fluorescence intensity imaging demonstrated that metabolically highly active cells and tissues, such as the ependyma, choroid plexus, or vascular endothelial cells, tended to show high signal intensity. When analyzed by fluorescence lifetime imaging, these structures also showed the longest lifetimes of endogenous fluorophores (>1700 ps) excited at 750 nm. In contrast, excitable fluorophores within erythrocytes were the shortest lived (900 ± 72 ps). In gray and white matter, the brain parenchyma showed an intermediate fluorescence lifetime (1380 ± 23 ps and 1360 ± 33 ps, respectively) (Fig. 3). Generally, the nuclei of glia showed low fluorescence intensity. The cytoplasm of glia cells frequently contained granules of high fluorescence intensity and relatively short fluorescence lifetime. Confirming our previous observations, the fluorescence lifetime of the nuclei increased with increasing excitation wavelengths (Leppert et al., 2006). The nuclei of hippocampal neurons also showed low fluorescence intensity (Fig. 4). The fluorescence lifetimes of endogenous fluorophores within hippocampal neurons showed a linear increase from 720 nm to 770 nm excitation, which was significantly different from the brain parenchyma neighboring the groups of neurons. These distinct excitation/lifetime profiles of cellular and subcellular structures reflect the photochemical composition of these regions of interest (Xu et al., 1996a, 1996b).
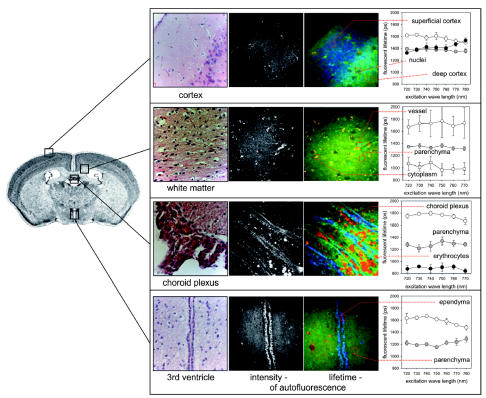
Fig. 3.
Microanatomical structures of normal mouse brain were analyzed by multiphoton excitation intensity imaging, color-coded fluorescence lifetime imaging (at 750 nm excitation), and conventional light microscopy of histological sections stained with hematoxylin and eosin. The excitation/lifetime profile of regions of interest within specific brain areas was calculated from at least three analyses (right panel). A representative region of interest for each anatomical site is illustrated by a framed area. Metabolically highly active cells of the ependyma, choroid plexus, and vascular endothelial cells showed high fluorescence on intensity imaging and tended to show long fluorescence lifetimes.
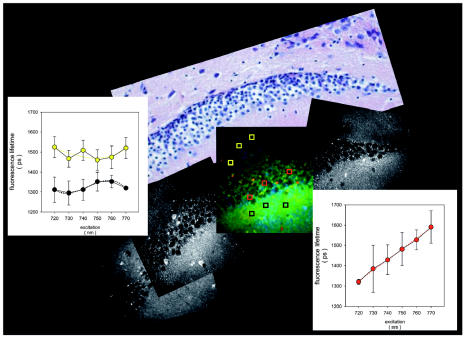
Fig. 4.
Fluorescence intensity images of hippocampal neurons in mouse brain. The inset shows the corresponding fluorescence lifetime image at an excitation wavelength of 750 nm. The graphs show the excitation/lifetime profiles for specific regions of interest (red, hippocampal neurons; yellow and black, adjacent parenchyma). The nuclei of the hippocampal neurons show a characteristic excitation/lifetime profile with a positive correlation of excitation wavelength to fluorescence lifetime.
Fluorescence Lifetime Analysis of Experimental Glioma Tissue
Obviously, multiphoton excitation microscopy and fluorescence lifetime imaging have the potential of providing cell-type-specific or tissue-specific information. We therefore used an intracranial tumor transplantation model in NMRI mice to study the relationship of fluorescence intensity and fluorescence lifetime of human-glioma-derived cells and the murine host brain.
Tumor-bearing mouse brains were obtained as described in Materials and Methods. Coronal sections (2 μm) were cut at the level of the implantation site, and the native tissue was subjected to multiphoton microscopy immediately. Intensity images allowed easy identification of the tumor transplants because of a profoundly increased signal intensity of the tumor cells at 750 nm excitation over normal cells of the white or gray matter and the surrounding brain parenchyma (Fig. 5A). Although our analysis of normal brain identified several highly autofluorescing cell types, these could be distinguished from tumor based on their distribution and specific morphology (compare Figs. 3 and 4). On intensity images, tumor-adjacent white matter showed a low density of low-signal-intensity nuclei. In contrast, the tumor transplants were highly cellular, with low-fluorescence-intensity nuclei and a high-signal-intensity cytoplasm. Continuous-spectrum color-coded fluorescence lifetime imaging of tumor and adjacent brain demonstrated that tumor tissue showed longer mean fluorescence lifetimes than did normal white matter or normal cortical gray matter (1780 ± 43 ps and 1540 ± 30 ps, respectively) (Fig. 5B and C). U87 cells implanted into mouse brain typically form a well-defined tumor-to-brain interface with few single invasive cells. Discrete color coding of lifetime ranges adapted to a region of interest in U87 tumors therefore resulted in an exact reproduction of the anatomical tumor-to-brain interface based on fluorescence lifetimes (Fig. 5D). Although fluorescence lifetimes differed among experimental tumors derived from the three cell lines that we studied, the fluorescence lifetime was always significantly longer (ranging from approximately 1640 to 1800 ps) than for tumor-adjacent brain (about 1510–1580 ps) at 750 nm excitation. The analysis of excitation/fluorescence lifetime profiles of U87 tumors and adjacent brain for increasing excitation wavelengths resulted in similar biphasic lifetime profiles for both tumor and adjacent brain (Fig. 6). However, the fluorescence lifetimes of tumor tissue were significantly longer at any excitation wavelength than those for brain tissue. A biphasic excitation/fluorescence lifetime profile with a maximum at approximately 750 nm excitation was observed in all experimental tumors derived from the three human glioma cell lines used (G-28, G-112, and U87) (data not shown). These findings are consistent with our previously reported excitation/fluorescence lifetime profiles of G-28, G-112, and U87 cells in monolayer culture (Leppert et al., 2006). Interestingly, the mean lifetimes of tumor-adjacent brain (1540 ps) at 750 nm excitation tended to be longer than the mean fluorescence lifetimes of brain more distant from the tumor or the mean lifetimes of white matter obtained from non-tumor-bearing animals, which generally were 1300–1400 ps. Fluorescence intensity images of tumor-adjacent brain and normal brain showed no difference.
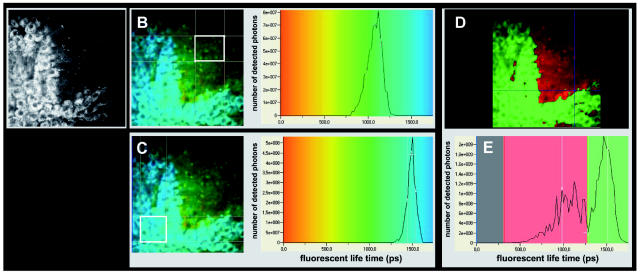
Fig. 5.
(A) Fluorescence intensity image of the tumor–brain interface in an experimental U87 glioma in NMRI mouse brain. (B and C) Continuous color-coded lifetime image of the tumor–mouse brain interface. (B) Histogram of the fluorescence lifetime distribution in a region of interest (white frame) corresponding to tumor-adjacent brain. (C) Fluorescence lifetime distributions in a region of interest corresponding to U87 glioma tissue, demonstrating the significantly longer fluorescence lifetimes of tumor tissue compared with adjacent mouse brain. (D) Two-color-coded image of the tumor–brain interface. The lifetime ranges for red and green were selected based on the peak distributions of the lifetime histogram. (E) Color-coded gating of fluorescence lifetimes allowed discrimination of tumor, and adjacent brain demonstrated a well-defined tumor–brain interface in U87 gliomas. All images shown here were obtained at an excitation wavelength of 750 nm.
Fig. 6.
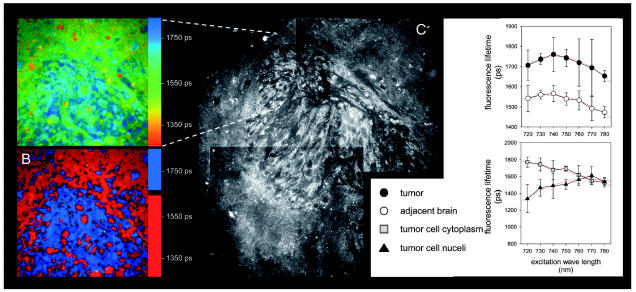
The continuous-spectrum color-coded fluorescence lifetime images of U87 glioma cells in mouse brain (A) and discrete color-coded spectrum for optimal discrimination of tumor and normal brain (B). Longer fluorescence lifetimes of tumor cells are coded blue. The intensity images show the microanatomy of the corresponding tumor area (C). Tumor cells showed higher fluorescence intensity and a prolonged fluorescence lifetime compared with surrounding brain. The analysis of individual tumor cells and adjacent brain showed that at all excitation wavelengths the fluorescence lifetimes exceeded those of normal brain parenchyma. The nuclei of tumor cells and cytoplasmic areas of tumor cells showed distinct excitation/lifetime profiles.
Multiphoton Excitation Autofluorescence Intensity and Lifetime Imaging of Glioma Biopsy Specimens
To determine whether fluorescence lifetime imaging could delineate adjacent brain and brain tumors in clinical specimens, tumor-adjacent brain and brain tumor biopsies were obtained at surgery and immediately subjected to multiphoton microscopy. Following the analysis, the tissues were fixed in formalin and processed for routine diagnostic histopathology.
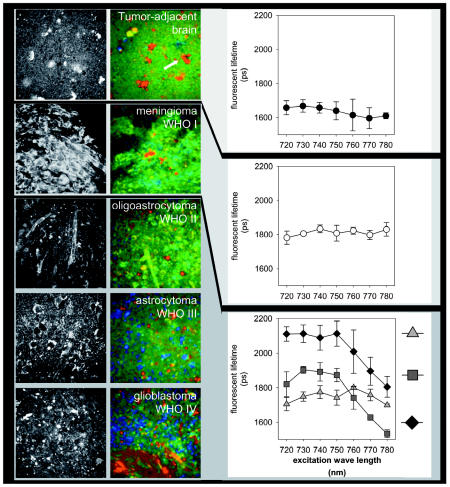
Tumor-adjacent human brain specimens showed structures similar to those in the murine brain specimens. Multiphoton intensity imaging demonstrated few nuclei per tissue volume and some vascular elements identified as capillaries. In contrast to normal mouse brain, human tumor-adjacent brain specimens contained a larger number of cells with granulated cytoplasm (Fig. 7). The intracellular granules were highly autofluorescing at an excitation wavelength of 750 nm. Fluorescence lifetime imaging showed very short-lived fluorophores within these intracellular compartments, with mean lifetimes of 560 ± 22 ps (white arrow, Fig. 7). Histologically, these cells corresponded to CD68+ macrophages (data not shown). The parenchyma of these brain specimens showed fluorescence lifetimes ranging from 1400 to 1750 ps at 750 nm excitation. A meningioma specimen showed high fluorescence intensity of the cytoplasm of tumor cells with low-signal-intensity nuclei. The excitation/fluorescence lifetime profile of the meningioma specimen showed a tendency to increased lifetimes at increased excitation wavelengths, but with generally longer lifetimes of endogenous fluorophores than for tumor-adjacent brain. We also analyzed three gliomas, a WHO grade II astrocytoma, an anaplastic astrocytoma, and a WHO grade IV glioblastoma. Fluorescence lifetime imaging at 750 nm demonstrated longer mean fluorescence lifetimes for the tumor parenchyma of all three glioma specimens than for tumor-adjacent brain. Strikingly, the glioblastoma specimen showed the longest mean lifetime of autofluorescence (2110 ± 73 ps), followed by the anaplastic glioma (1870 ± 40 ps) and the low-grade astrocytoma (1740 ± 44 ps). In these specimens, the excitation/fluorescence lifetime profiles demonstrated that, in both the anaplastic astrocytoma and the glioblastoma, the fluorescence lifetimes were decreased at excitation wavelengths greater than 750 nm (Fig. 7).
Fig. 7.
Multiphoton excitation fluorescence intensity imaging and fluorescence lifetime imaging (at 750 nm) of human tumor-adjacent brain and specimens of human brain tumors. For fluorescence lifetime imaging of the cytoplasmic area of tumor cells, identical parameters of continuous-spectrum color coding were used for all specimens. The analysis of fluorescence lifetimes at increasing excitation wavelengths demonstrated distinct excitation/lifetime profiles for normal brain and malignant gliomas.
Discussion
Near-infrared multiphoton excitation laser scanning microscopes can potentially be employed as novel non-invasive biomedical tools for three-dimensionally and time-resolved imaging of fluorophores in optical tissue diagnostics. This includes structural imaging at the sub-cellular level, as well as the photochemical characterization of living tissues and functional imaging of solid tissues.
The first biomedical applications in experimental dermatology have already demonstrated the potential of this technology, and the DermaInspect multiphoton microscope used in this study is now commercially available for clinical applications in dermatology (König and Rieman, 2003). We have recently applied this technology to structural and photochemical imaging of cultured glioma cells and experimental gliomas ex vivo. This conceptual study demonstrated that high microanatomical definition of the tumor parenchyma, invasion zone, and normal adjacent brain can be obtained in unprocessed tissue blocks. In an intracranial mouse model, fluorescence intensity images allowed delineation of single tumor cells invading murine brain (Leppert et al., 2006). However, the characterization of individual cells was not limited to structural anatomical imaging. Fluorescence lifetime imaging of cultured glioma cells in vivo demonstrated that subcellular compartments showed different excitability and fluorescence lifetimes of endogenous fluorophores. Among the fluorophores for which fluorescence lifetime spectra have already been characterized are NADPH, flavines, lipofuscin, elastin, collagen, and melanin (König and Rieman, 2003). However, no studies have investigated specific fluorophores in the brain.
Fluorescence lifetime images allowed delineation of the peripheral and perinuclear cytoplasm, intracellular granules, and the nucleus. Owing to the limited number of animals used in this study, we were not able to give quantitative values for the sensitivity and specificity. The aim of this study was to show that we can detect spectroscopic differences (characteristic dependence of the fluorescence lifetime on the excitation wavelength). The hardware-dependent limitation of the time resolution, as described in Materials and Methods, were found to be errors of up to 10% of the measured lifetimes. Typically, the differences in the lifetimes of the different tissue components were found to be 20%–50%. Therefore, at least 200 photons have to be detected to separate cell types by two standard deviations of the measured decay time. With a typical count rate of 50,000 photons/s, 250 pixels/s can be measured.
The analysis of the relationship between the laser excitation wavelength and the lifetime of endogenous fluorophores showed characteristic profiles for intracellular compartments in cultured glioma cells. Interestingly, these excitation/fluorescence lifetime profiles of cultured glioma cell lines and primary cultures of gliomas differed from profiles obtained from cultured cells derived from other tumor types (Leppert et al., 2006). This suggests that fluorescence lifetime spectroscopy may differentiate histotypes of cells based on the excitability of cell-type-specific expression of endogenous chromophores, their chemical states, or their interaction with other biomolecules. Whether fluorescence lifetime spectroscopy may be extended to a discrimination of functional cellular states in cultured glioma cells is currently under investigation.
Our aim in the present study was to analyze whether fluorescence lifetime imaging may be able to identify glioma cells in situ. Such analysis requires the characterization of fluorescence intensities and fluorescence lifetimes within normal brain and brain tumors. Therefore, we used an intracranial model system in NMRI mice using transplantable human glioma cell lines (Brockmann et al., 2006). This study demonstrated that multiphoton microscopy in native tissue blocks allows a detailed display of microanatomical brain structures without the need of contrasting techniques.
Fluorescence lifetime imaging showed that gray and white matter represent areas of homogeneous distribution of fluorescence lifetimes at any excitation wavelength between 720 and 770 nm. Per tissue volume, few nuclei and cytoplasmic structures of resident cells could be identified. Within this homogeneous background, the tumor transplants could be readily identified, because of high fluorescence intensity of the cytoplasmic areas of tumor cells, which contrasted with the low-intensity nuclei. At any excitation wavelength, the tumors derived from the three different cell lines showed markedly longer fluorescence lifetimes than did adjacent gray or white matter.
Interestingly, the fluorescence lifetime of tumor-adjacent brain was consistently longer than that of normal white matter. Whether this is a consequence of tissue edema or ingress of cellular elements responding to the tumor stimulus remains open. However, several microanatomical structures were identified within normal brain that by multiphoton excitation showed intense autofluorescence and long fluorescence lifetimes of excited fluorophores. The lifetimes of the ventricular ependyma, for example, reached values similar to those for tumor cells. Interestingly, the elements of normal brain showing long fluorescence lifetimes were composed of metabolically highly active cell types. Further examples were cells of the epithelium of the choroid plexus and endothelial cells of capillaries and larger blood vessels. Because of their specific morphology, these anatomical structures and normal cells could be easily distinguished from tumor.
In the deep basal ganglia, however, single cells of high fluorescence intensity and long fluorescence lifetime were observed in normal brain specimens (compare Fig. 2). These individual cells showed excitation spectra similar to those of tumor transplants. This may suggest, on the basis of the parameters analyzed here, that multiphoton excitation fluorescence intensity imaging and fluorescence lifetime spectroscopy offer no tumor specificity but rather may identify metabolically highly active tissues. This is further supported by a recent observation (using similar detection parameters) that the intestinal endothelium shows intense autofluorescence and long fluorescence lifetimes (Gebert et al., manuscript in preparation).
Nevertheless, fluorescence lifetime imaging of malignant human glioma specimens showed that the fluorescence lifetimes and the excitation/lifetime profiles of tumor specimens were significantly different from those of tumor-adjacent brain. These data would suggest that multiphoton excitation of autofluorescence theoretically provides means for a tissue analysis in situ, which could be used, for example, to detect residual tumor at the resection edge. In contrast to conventional one-photon laser scanning microscopy, femtosecond pulsed laser microscopy of living specimens can be performed at peak intensities of 200 GW/cm2 with no sign of structural or functional photodamage. This has been demonstrated for cells in monolayer culture as well as for mammalian embryos and human skin (Masters et al., 1997, 1998; Oehring et al., 2000; Squirrell et al., 1999; Tyrell and Keyse, 1990). Therefore, intraoperative in vivo multiphoton microscopy of brain tissue conceivably could provide a high-resolution, noninvasive diagnostic tool. Recent developments of this technology have introduced miniaturized scanner probes connected to optic fibers that have been used for in vivo imaging of the mouse central nervous system over extended periods of time (Kim et al., 2004). Such probes placed in direct contact with the target tissue may offer future solutions to high-resolution optical imaging of a target volume that follows the respiratory and arterial cycle, such as the brain.
Footnotes
This study was supported by grants of the University Hospital of Schleswig-Holstein, Campus Lübeck (A.G., J.L., and N.P.), the Kreitz Foundation (J.L. and A.G.), and the Future Investment Program of Schleswig-Holstein and the Deutsche Forschungsgemeinschaft (A.G. and G.H.).
References
- Becker W, Bergmann A, König K, Tirlapur U. Picosecond fluorescence lifetime microscopy by TCSP imaging. Proc SPIE. 2001;4262:414–419. [Google Scholar]
- Brockmann MA, Ulmer S, Leppert J, Nadrowitz R, Wuestenberg R, Nolte I, Petersen D, Groden C, Giese A, Gottschalk S. Analysis of mouse brain using a clinical 1.5 tesla scanner and a standard small loop surface coil. Brain Res. 2006;1068:138–142. doi: 10.1016/j.brainres.2005.10.098. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim KH, Yazdanfar S, So PTC. High-speed hand-held multiphoton multifoci microscopy. Proc SPIE. 2004;5323:267–272. [Google Scholar]
- Köllner M, Wolfrum J. How many photons are necessary for fluorescence-lifetime measurements? Chem Phys Lett. 1992;200:199. [Google Scholar]
- König K. Multiphoton microscopy in life science. J Microsc. 2000;200:83–104. doi: 10.1046/j.1365-2818.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- König K, Riemann I. High-resolution multiphoton tomography of human skin with subcellular spatial resolution and picosecond time resolution. J Biomed Opt. 2003;8:432–439. doi: 10.1117/1.1577349. [DOI] [PubMed] [Google Scholar]
- König K, Schenke-Layland K, Riemann I, Stock UA. Multiphoton autofluorescence imaging of intratissue elastic fibers. Biomaterials. 2005;26:495–500. doi: 10.1016/j.biomaterials.2004.02.059. [DOI] [PubMed] [Google Scholar]
- Leppert J, Krajewski J, Kantelhardt SR, Schlaffer S, Petkus N, Reusche E, Huttmann G, Giese A. Multiphoton excitation fluorescence microscopy of glioma tissue. Neurosurgery. 2006;58:759–767. doi: 10.1227/01.NEU.0000204885.45644.22. [DOI] [PubMed] [Google Scholar]
- Masters BR, So PT, Gratton E. Multiphoton excitation fluorescence microscopy and spectroscopy of in vivo human skin. Biophys J. 1997;72:2405–2412. doi: 10.1016/S0006-3495(97)78886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters BR, So PT, Gratton E. Multiphoton excitation microscopy of in vivo human skin. Functional and morphological optical biopsy based on three-dimensional imaging, lifetime measurements and fluorescence spectroscopy. Ann N Y Acad Sci. 1998;838:58–67. doi: 10.1111/j.1749-6632.1998.tb08187.x. [DOI] [PubMed] [Google Scholar]
- Oehring H, Riemann I, Fischer P, Halbhuber KJ, König K. Ultrastructure and reproduction behaviour of single CHO-K1 cells exposed to near infrared femtosecond laser pulses. Scanning. 2000;22:263–270. doi: 10.1002/sca.4950220406. [DOI] [PubMed] [Google Scholar]
- Squirrell JM, Wokosin DL, White JG, Bavister BD. Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability. Nat Biotechnol. 1999;17:763–767. doi: 10.1038/11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teuchner K, Freyer W, Leupold D, Volkmer A, Birch DJ, Altmeyer P, Stucker M, Hoffmann K. Femtosecond two-photon excited fluorescence of melanin. Photochem Photobiol. 1999;70:146–151. [PubMed] [Google Scholar]
- Tyrell RM, Keyse SM. New trends in photobiology: The interaction of UVA radiation with cultured cells. J Photochem Photobiol. 1990;4:349–361. doi: 10.1016/1011-1344(90)85014-n. [DOI] [PubMed] [Google Scholar]
- Xu C, Williams RM, Zipfel W, Webb WW. Multiphoton excitation cross-sections of molecular fluorophores. Bioimaging. 1996a;4:198–207. [Google Scholar]
- Xu C, Zipfel W, Shear JB, Williams RM, Webb WW. Multiphoton fluorescence excitation: New spectral windows for biological nonlinear microscopy. Proc Natl Acad Sci U S A. 1996b;93:10763–10768. doi: 10.1073/pnas.93.20.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]