Abstract
The receptor protein tyrosine phosphatase beta (RPTPβ/PTPζ) is overexpressed in glioblastoma tumors and plays a functional role in tumor cell migration and adhesion. Glioblastomas express at least three splice variants of RPTPβ, including long and short receptor forms and a secreted chondroitin sulfate proteoglycan called phosphacan. Here we explore the differences in the expression pattern and function of long RPTPβ and short RPTPβ. The short form of RPTPβ lacks exon 12, which encodes 860 amino acids located in the extracellular domain. Until now, functional differences between long and short RPTPβ have been difficult to elucidate. In this study, antibodies specific to the splice junction, unique to short RPTPβ, allowed for the discrimination of the two receptors. A study of normal brain tissue and graded astrocytomas indicates that long and short RPTPβ forms have an overlapping expression pattern. In order to study functional differences between long and short RPTPβ, we created stable U87 glioblastoma cells that expressed these receptors. U87 stable cell lines overexpressing long or short RPTPβ migrate faster and adhere more robustly than parental U87 cells. The two forms differ in that long-RPTPβ -overexpressing cells migrate and adhere better than short-RPTPβ -overexpressing cells. A study of the extracellular domain of short RPTPβ indicates that it retains much of the functional capacity of phosphacan. Indeed, the action of recombinant, short-RPTPβ extra-cellular domain protein is similar to that of phosphacan as a repulsive substrate for glioblastoma cells. Comparison of the signaling capacity of long RPTPβ to that of short RPTPβ reveals very similar abilities to activate transcription pathways. Moreover, transient transfection with either long or short RPTPβ activates NF-κB reporter gene transcription. Because of their tumor-restricted and largely overlapping expression patterns in glioblastoma, both RPTPβ splice forms are potential therapeutic targets. The involvement of long and short RPTPβ in glioma tumor cell biology also contributes to the value of RPTPβ as a cancer target.
Receptor protein tyrosine phosphatase beta (RPTPβ),2 also known as protein tyrosine phosphatase zeta (PTPζ), is a member of a diverse class of PTPs consisting of both cytoplasmic PTPs and transmembrane, receptor-like PTPs (e.g., RPTPβ). Cellular tyrosine phosphorylation plays a crucial role in the control of growth, migration, differentiation, and transformation of eukaryotic cells and is regulated by the balanced action of protein tyrosine kinases and protein tyrosine phosphatases. Whereas most PTPs are expressed in peripheral tissues and the CNS, RPTPβ expression is restricted to the CNS (Levy et al., 1993). Interestingly, the highest levels of RPTPβ expression in the developing mouse brain are in regions that have the greatest mitotic potential, that is, the embryonic ventricular and subventricular zones, the dentate gyrus, and the subependymal layer of the anterior horn of the lateral ventricle (Levy et al., 1993). The expression profile of RPTPβ is consistent with its suggested role in cell adhesion, migration, and invasion. In normal settings, RPTPβ and phosphacan seem to facilitate the development and maintenance of the CNS (Milev et al., 1994). In the setting of glioma, RPTPβ is preferentially expressed in these tumors and likely contributes to increased malignancy (Mueller et al., 2004; Ulbricht et al., 2003).
The function of RPTPβ is regulated in part by the expression pattern of its various isoforms and by the availability of ligands to bind to the extracellular domain. Three splice variants of RPTPβ have been described: a full-length 9.4-kb transmembrane transcript, a short 6.4-kb transmembrane transcript, and a soluble 8.4-kb transcript, known as 6B4 proteoglycan/phosphacan (Levy et al., 1993). More recently, a novel truncated form of phosphacan (phosphacan short isoform) has been identified that contributes to cell-cell and cell–extracellular matrix interactions (Garwood et al., 2003). Various cell adhesion molecules (e.g., NgCAM, Nr-CAM, and contactin) and extracellular matrix molecules (e.g., tenascin-C) are known to bind to RPTPβ (Table 1). The extracellular domain of RPTPβ has three identifiable domains, the carbonic anhydrase (CAH), fibronectin III (FNIII), and glycine-serine-rich regions. These domains and glycosylation contribute to the interaction of RPTPβ with its various ligands (see references in Table 1). We tested the functional importance of these domains by comparing cells expressing long and short RPTPβ in response to various ligands. Pleiotrophin (PTN), the first identified soluble ligand of RPTPβ, inactivates the phosphatase activity of RPTPβ. This loss of activity in turn leads to increased tyrosine phosphorylation of β -catenin (Meng et al., 2000). Little else is known about the signal transduction capacity of the RPTPβ splice forms. Therefore, we explored the relative contribution of the long and short forms of RPTPβ to signal transduction. In this study, our results establish that both transmembrane forms of RPTPβ are overexpressed in glioblastoma. In addition, we demonstrate that both long RPTPβ and short RPTPβ play an important role in the regulation of glioma cell migration and adhesion in response to various ligands. The characterization of long-RPTPβ and short-RPTPβ signaling is important to the development of functional markers and therapeutics for the analysis and treatment of glioma.
Table 1.
RPTPβ extracellular domain binding partners
| Binding Partner | Region of RPTPβ | Reference |
|---|---|---|
| Contactin | CAH domain | Peles et al., 1998 |
| Tenascin-C | FNIII domain | Barnea et al., 1994 |
| Grumet et al., 1994 | ||
| Adamsky et al., 2001 | ||
| Ng-CAM | CAH domain | Milev et al., 1994 |
| Nr-CAM | CAH domain | Sakurai et al., 1996 |
| Pleiotrophin | Low- and high- affinity sites | Maeda et al., 1996 |
| Meng et al., 2000 |
Abbreviations: CAH, carbonic anhydrase; CAM, cell adhesion molecule; FNIII, fibronectin III.
Experimental Procedures
Materials
Human U87-MG (American Type Culture Collection cell line) and U373 (European Collection of Cell Cultures) cell lines were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Gibco, Invitrogen Corporation, Carlsbad, Calif.), 1 mmol/liter l-glutamine, and 100 U/ml penicillin/streptomycin. Mouse monoclonal antibodies against the carboxyl-terminus of RPTPβ were obtained from Transduction Labs (BD Biosciences, San Jose, Calif.). Custom rabbit polyclonal antibodies directed against RPTPβ splice junction (CTQPVYNEASNSSHES) were obtained from Zymed Laboratories, Inc. (South San Francisco, Calif.). Secondary anti-rabbit-horseradish peroxidase (HRP) and anti-mouse-HRP conjugates were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Alexa 594 phalloidin was obtained from Molecular Probes (Eugene, Oreg.). Ligands and extra-cellular matrix components were prepared according to manufacturer’s guidelines and as described in the adhesion and migration assays. PTN and Collagen I were purchased from Sigma (St. Louis, Mo.). Tenascin-C and the cell adhesion molecule NgCAM were purchased from Chemicon (Temecula, Calif.).
Immunohistochemistry
Ambion LandMark Tissue MicroArray low-density brain slides (Ambion, Austin, Tex.) were used to study the expression profiles of long and short RPTPβ. Slides were placed on a heat block at 45ºC for 4 to 6 h and dewaxed with EZ-DeWax solution (Innogenex of Bio-genex, San Ramon, Calif.). Slides were then placed in a bath containing target retrieval solution (Retrievit, Innogenex) and simmered for 15 min in a microwave. Slides were stained with either the carboxyl-terminus RPTPβ antibody (Transduction Labs) or the custom rabbit polyclonal antibodies specific to the short-RPTPβ junction (Zymed). The slides were processed by using either anti-mouse or anti-rabbit immunohistochemistry kits with DAB (diaminobenzidine) colorimetric end-point detection and hemotoxylin counterstain (Innogenex).
Immunoblot Analysis
Cells were lysed in radioimmunoprecipitation assay buffer (0.1% sodium dodecyl sulfate [SDS], 1% NP40, and 0.5% sodium deoxycholate in phosphate-buffered saline [PBS]) for 20 min on ice and then cleared by centrifugation. Total cell lysate protein was quantified by using BCA Protein Assay Reagent (Pierce, Rockford, Ill.). Cellular proteins were separated on 8% SDS-polyacrylamide gels (Invitrogen) and blotted onto nitro-cellulose membranes. Membranes were next blocked in 5% nonfat milk and then incubated with the indicated antibody. After incubation with an HRP-conjugated secondary antibody, immunodetection was accomplished by an enhanced chemiluminescence method (Amersham, Piscataway, N.J.), followed by exposure to X-ray film (Kodak, Rochester, N.Y.).
Stable Cell Lines
Stable cell lines were created by transfecting U87 cells with 2 μg of either pcDNA3.1, pcDNA3.1-long-RPTPβ, or pcDNA3.1-short-RPTPβ, by using the FuGene 6 transfection reagent according to manufacturer’s instructions (Roche, Indianapolis, Ind.). Stable cell lines were selected by incubating cells with medium containing 500 μg/ml of G418 (Gibco). Surviving cells were pooled and then expanded and characterized.
Migration Assay
Human U87 cells were seeded into the upper chamber of 24-well Fluoroblok migration plates (Becton Dickinson and Company, Franklin Lakes, N.J.) at 25,000 cells per well in serum-free medium. The lower wells contained 5% fetal bovine serum medium. The migration chambers were coated with10 μg/ml PTN, tenascin-C, NgCAM, or Collagen I for 1 h at room temperature, followed by bovine serum albumin (BSA) blocking. The plates were incubated for 4 h at 37ºC with 5% CO2. The migration chambers were then stained for live cells by using calcein AM (Molecular Probes), and the fluorescence was measured on a plate reader.
Adhesion Assay
To measure adhesion of cells onto coated surfaces, 96-well plates were incubated for 1 h at room temperature with 10 μg/ml BSA, chondroitin sulfate proteoglycan (phosphacan), PTN, tenascin-C, NgCAM, or Collagen I. The plates were then washed with PBS, blocked for 1 h at room temperature with 1% BSA, and washed again. Harvested cells were suspended into serum-free media and seeded onto the coated plates at a density of 30,000 cells per well. Following incubation for 30 min at 37°C with 5% CO2, the extent of adhesion was measured. The plates were washed three times with PBS, and the number of viable, adherent cells was measured by using CellTiter-Glo reagent according to manufacturer’s instructions (Promega, San Luis Obispo, Calif.).
RPTPβ Protein Expression and Purification
The extracellular domain (ECD) for short RPTPβ (residues 26–774), including the unique splice junction, was expressed with a C-terminus 6xHis tag by using baculovirus. Briefly, PCR was employed to introduce the codons for the 6xHis tag to the gene sequence encoding the ECD, along with appropriate restriction sites. The construct was subcloned into pFastBac1 (Invitrogen) and transformed into DH10Bac cells (Invitrogen) for bacmid recombination. Recombinant bacmid was isolated and transfected into Sf9 cells for baculovirus generation. Recombinant baculovirus was amplified and used to infect Sf9 cells for protein production. The native secretion signal for short RPTPβ ECD was utilized by the insect cells to secrete the protein into serum-free media (Expression Systems, Woodland, Calif.). The protein was purified from the media with two chromatography steps: immobilized metal affinity (Ni2+-NTA FF, Qiagen, Valencia, Calif.) followed by anion exchange (Q Sepharose FF, Amersham Biosciences).
Reporter Assay
HEK 293 cells were seeded at 10,000 cells per well in a 96-well plate. The next day quadruplicate wells of cells were cotransfected by using FuGene transfection reagent (Roche) with 50 ng of NF-κB-Luc, along with 10 ng of RSV-β -galactosidase as a normalization control, and either 100 ng of pcDNA3.1, 100 ng pcDNA3.1-long-RPTPβ, or 100 ng pcDNA3.1-short-RPTPβ. Twenty-four hours after transfection, the cells were lysed and assayed for luciferase and β -galactosidase activity by using the Tropix Dual-Light Reporter Kit (Applied Bio-systems, Bedford, Mass.). All results were normalized to β -galactosidase and are representative of three independent experiments.
Results
Comparison of Long and Short RPTPb Forms
The RPTPβ transcript can be spliced into long and short receptor forms and a third secreted protein lacking catalytic activity, phosphacan (Fig. 1A). The ECD of these proteins is characterized by a CAH domain, an FNIII domain, and several chondroitin sulfate–glycosaminoglycan sites located in a large glycine-serine-rich domain. Demonstrated RPTPβ ligands (contactin, tenascin-C, NgCAM, Nr-CAM, PTN, and others) interact with these domains by various means. Table 1 summarizes some previously reported mapping of the binding partner with the region of RPTPβ or phosphacan. Comparing the differences between long and short RPTPβ with regard to expression and function is the focus of our studies, and we relate here the known RPTPβ interactions with functional differences between the long and short receptor isoforms.
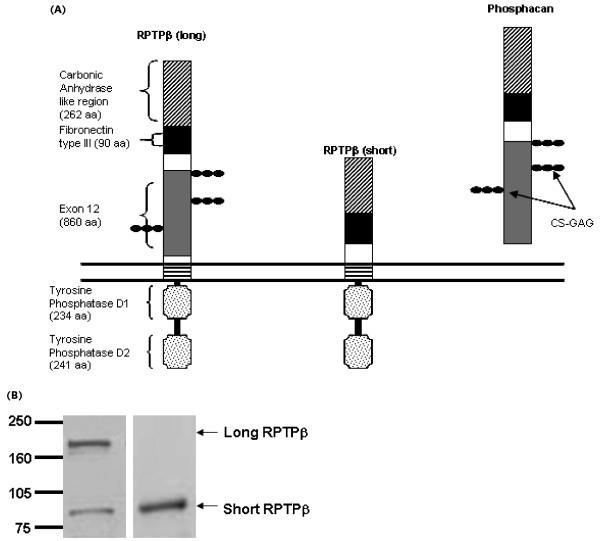
Fig. 1.
Domain organization of long RPTPβ, short RPTPβ, and phosphacan. (A) All three proteins have carbonic anhydrase (CAH) and fibronectin type III domains. Short RPTPβ lacks the bulk of the glycine-serine-rich domain encoded by exon 12. This exon also encodes sites for chondroitin sulfate–glycosaminoglycan moieties and is present in both long RPTPβ and phosphacan. Long and short RPTPβ have a short juxtamembrane region, and in short RPTPβ a unique, in-frame junction is formed in the absence of the exon 12–encoded domain. (B) Expression of both long and short RPTPβ in U373 glioblastoma. The left panel shows an immunoblot probed with RPTPβ carboxyl-terminus antibody, recognizing the intracellular region of both long and short RPTPβ. The right panel shows an immunoblot, from the same lysate, probed with antibodies specific to the short RPTPβ protein.
Unlike long RPTPβ, short RPTPβ lacks the 860 amino acids comprising the bulk of the glycine-serine-rich domain (encoded by exon 12). This splice event generates a new junction site in the ECD that is unique to short RPTPβ. In order to study this unique feature of short RPTPβ, we generated rabbit polyclonal antibodies against a peptide that maps to the splice junction found in short RPTPβ. These antibodies should specifically recognize short RPTPβ but not long RPTPβ or phosphacan. U373 cells, which have been demonstrated to express both long and short RPTPβ as well as phosphacan (Meng et al., 2000) were used to test the specificity of the generated antibody. Fig. 1B (left panel) shows that both long and short RPTPβ are detected in U373 cells with a carboxyl-terminus antibody, but only short RPTPβ is recognized by the junction-specific antibodies (right panel). Using the carboxyl-terminus antibody in conjunction with the short RPTPβ splice junction–specific antibodies provides a means of distinguishing between long and short RPTPβ.
Expression Patterns of Long and Short RPTPß Forms Broadly Overlap
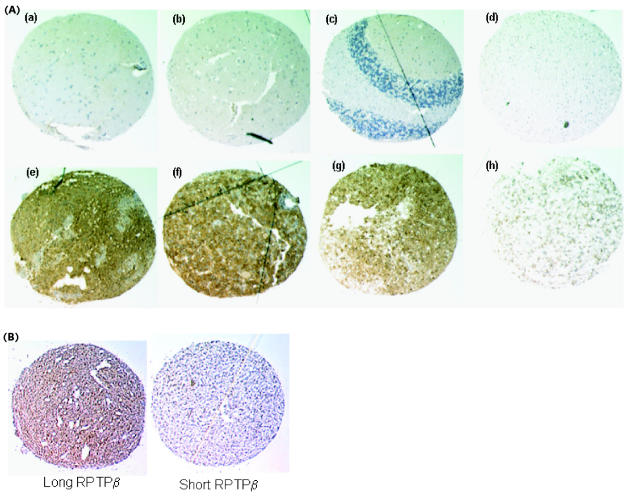
In order to compare long and short RPTPβ expression patterns we used immunohistochemistry to analyze a collection of variably graded astrocytomas along with normal brain tissue. In this study, 16 of 24 glioblastoma tumors (66%) stained positive for RPTPβ when we used the carboxyl-terminus-directed antibody that recognizes both long and short receptor forms but not phosphacan (Fig. 2 and Table 2). This analysis expands upon the existing knowledge of RPTPβ expression in astrocytoma and normal adult human brain (Mueller et al., 2003; Ulbricht et al., 2003). Additional sections from the same tissue specimens were stained for short RPTPβ by using the junction-specific antibodies. The expression of short RPTPβ coincided with that of long RPTPβ. In fact, only one of the glioblastoma specimens expressing long RPTPβ did not express abundant levels of short RPTPβ (Fig. 2B). Differences in signal intensity between the carboxyl-terminus RPTPβ antibody and the short RPTPβ junction–specific antibody were evident. However, conclusions cannot be drawn from this observation because of the inherent differences that can exist between any two given antibodies and their abilities to work in such a technique. Therefore, these studies simply provide evidence that both long- and short RPTPβ are expressed in glioblastoma. Because long RPTPβ and phosphacan have nearly identical ECDs, this technique cannot distinguish these two forms, given the existing ECD antibodies. These results are consistent with our Northern blot and Western blot analysis (Mueller et al. [2003] and data not shown) and demonstrate the upregulation of long and short RPTPβ protein levels in glioblastoma with low to undetectable levels of expression in normal brain tissue.
Fig. 2.
Immunohistochemical analyses of normal brain and astrocytoma tissue demonstrate tumor-specific expression of RPTPβ. (A) Sections from normal brain (a–d), glioblastoma (e–g), and astrocytoma grade III tissue (h) were stained with RPTPβ carboxyl-terminus antibody (brown DAB chromagen) and counterstained with hemotoxylin (blue). Representative samples of normal brain sections (including cerebellum [c] and RPTPβ -positive glioblastoma tumors) are shown here. (B) Glioblastoma tissue sections. Of the 24 glioblastoma tissues tested in this matched set of sections, only one had significant stain for long RPTPβ (left panel) but did not stain for short RPTPβ (right panel).
Table 2.
Immunohistochemistry of RPTPβ expression
| Tumor Type | Incidence | Average Area | Average Intensity (0–3) |
|---|---|---|---|
| Normal brain | 0 of 4 (0%) | 0% | 0 |
| Glioblastoma | 16 of 24 (66%) | 65% | 2 |
| Astrocytoma III | 1 of 3 (33%) | 50% | 2 |
| Astrocytoma II | 3 of 4 (75%) | 100% | 1.5 |
Stable U87 Cell Lines Expressing Long or Short RPTPß Display Measurable Differences in Function
In order to study any functional differences between long and short RPTPβ, we generated full-length cDNA constructs encoding the two receptor variants. These constructs were used to generate stable U87 cell lines overexpressing either the long or short forms of RPTPβ (Fig. 3A). The parental U87 cells and the long-RPTPβ - and short-RPTPβ -expressing U87 cells were lysed and immunoprecipitated by using anti-carboxyl-terminus RPTPβ antibody. The immunoprecipitates were then blotted with the same antibody in order to measure the relative expression levels of transfected long and short RPTPβ forms, as well as endogenous receptors in the cell lines.
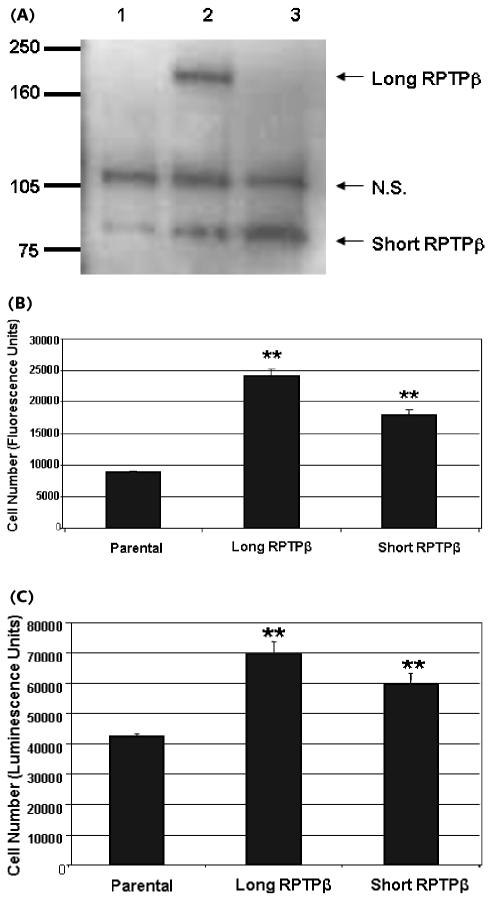
Fig. 3.
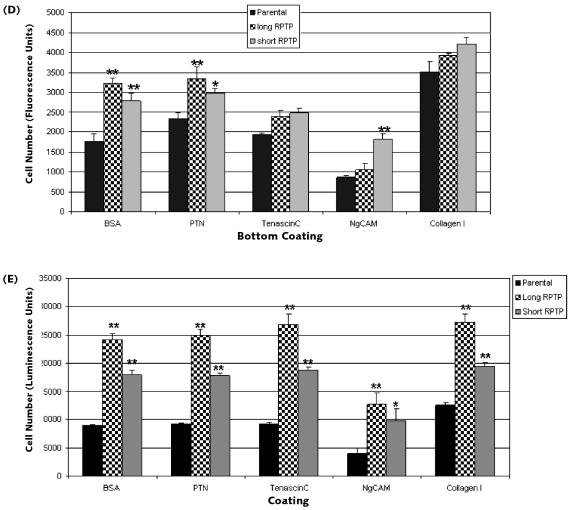
Comparison of U87 stable lines overexpressing long and short RPTPβ for their ability to migrate and adhere. (A) Parental (lane 1), long RPTPβ (lane 2), and short RPTPβ (lane 3) U87 cells were lysed, immunoprecipitated, and blotted by using anti-carboxyl-terminus RPTPβ antibody. Arrows indicate the expression of short and long RPTPβ and a nonspecific band (N.S.). (B) Parental U87 cells, cells overexpressing long RPTPβ, or cells overexpressing short RPTPβ were plated onto the upper chamber of a Fluoroblok migration plate. The cells were incubated for 4 h, and the cells that migrated were stained with calcein AM and detected by using a fluorescent plate reader. Both long- and short-RPTPβ -overexpressing cells migrate better than the parental line (error bars, ±SD; **P < 0.005). (C) Parental U87 cells, cells overexpressing long RPTPβ, or cells overexpressing short RPTPβ were plated and allowed to attach for 30 min. Following three washes with PBS, the number of viable adherent cells was measured. U87 stable lines expressing either long RPTPβ or short RPTPβ are more adherent than parental cells (error bars, ±SD; **P < 0.005). (D) Parental U87 cells, cells overexpressing long RPTPβ, or cells overexpressing short RPTPβ were plated onto the upper chamber of a Fluoroblok migration plate that had been coated on the lower side of the migration surface with the indicated ligands/substrates. Both long- and short-RPTPβ -overexpressing cells migrated moderately better than the parental line. However, long- and short- RPTPβ cells migrated similarly on most substrates except for NgCAM (error bars, ±SD; **P < 0.005; *P < 0.05). (E) Parental U87 cells, cells overexpressing long RPTPβ, or cells overexpressing short RPTPβ were plated and allowed to attach on plates coated with the indicated ligands/substrates for 30 min. U87 stable lines expressing either long RPTPβ or short RPTPβ were more adherent than parental cells on these substrates. However, long-RPTPβ -overexpressing cells adhered significantly better, even on the nonligand substrate Collagen I (error bars, ±SD; **P < 0.005; *P < 0.05).
These stable cell lines were used for subsequent functional studies. The stable cell lines were tested for the ability to migrate in a Fluoroblok (Becton Dickinson) migration chamber assay. Equal numbers of cells from each stable line were plated and allowed to migrate toward serum-containing media. Cells that migrated across the 8-μm porous membrane during the 4-h experiment were stained with calcein AM and visualized on a fluorometer. U87 cells overexpressing long RPTPβ migrated at higher numbers (33% higher) than cells overexpressing short RPTPβ (Fig. 3B). Both long-RPTPβ - and short-RPTPβ -overexpressing cells migrated faster than the parental control cells (167% and 100% increase, respectively).
Cell migration depends on coordinated cell attachment, detachment, and movement. Once a cell has attached to a surface, it can then flatten and extend processes that facilitate its coordinated detachment and movement. The attachment component of cell migration was measured for U87 cells overexpressing long and short RPTPβ by using a cell adhesion assay. U87 cells overexpressing either long or short RPTPβ were plated and allowed to adhere for 30 min. The plates were then washed to remove unattached cells, and the number of remaining cells on the plates was measured. Cells over-expressing long or short RPTPβ adhered better than parental control cells (65% and 40% more adherent cells, respectively) (Fig. 3C). These results agree with the migration data, indicating that long RPTPβ has a consistently greater effect than short RPTPβ in both cell migration and adhesion.
Various cell adhesion molecules (e.g., NgCAM, NrCAM, and contactin) and extracellular matrix molecules (e.g., tenascin-C) are known to bind to the extra-cellular domain of RPTPβ. PTN is the primary soluble factor demonstrated to bind to RPTPβ. In order to test the responses of long- and short-RPTPβ -expressing cells to their identified ligands, we examined cell migration and adhesion. In the migration experiments, the lower surface of the migration chambers was coated with BSA and Collagen I as controls, as well as PTN, tenascin-C, and NgCAM, as test conditions. The cell lines were then plated and allowed to migrate in the presence of these ligands (Fig. 3D). Collagen I coating facilitated robust migration for all three cell lines irrespective of RPTPβ status. Both long- and short-RPTPβ cells migrated more robustly than the parental line on BSA coating (considered our baseline). Long and short RPTPβ migrated similarly to each other in the presence of most specific substrates. Both long- and short-RPTPβ cells migrated better than parental in response to PTN. Tenascin-C had little effect on migration under the conditions we used. NgCAM, which was generally repulsive in this context, did not seem to affect short-RPTPβ -expressing cells when compared to parental cells. The RPTPβ ligands have also been studied for their role in cell adhesion. Therefore, we set out to examine the differences between long and short RPTPβ during cell attachment.
In a design similar to that detailed above, the cell lines were tested for their ability to adhere to surfaces coated with BSA, PTN, tenascin-C, NgCAM, and Collagen I. Consistent with previous experiments, both long- and short-RPTPβ cells adhered better than the parental line, wherein long RPTPβ attached more effectively than the other lines (Fig. 3E). Interestingly, long- and short-RPTPβ cells attached significantly better than the parental line onto Collagen I. In most cases, modest but measurable ligand dependency was observed between long- and short-RPTPβ cells when exposed to these substrates. The adhesion assays revealed greater differences between long and short RPTPβ than the migration assay. This is consistent with the observation that both long and short RPTPβ share the main functional domains (CAH and FNIII), and the established role of these ligands is in cell adhesion/repulsion.
Extracellular Domain of Short RPTPß Acts as Repulsive Substrate During Cell Adhesion
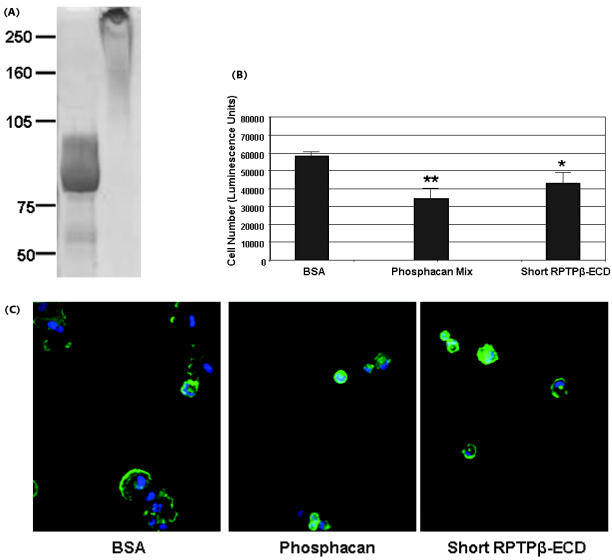
Most of the sites of interaction between RPTPβ and its ligands appear to map to the CAH and FNIII domains (Table 1). Little is known about the role of the glycine-serine-rich domain or the role that short RPTPβ plays in cell functions. In order to test the functional role of short RPTPβ in cell adhesion, we explored the possibility that the ECD of short RPTPβ could act to modulate glioblastoma cell adhesion. To this end, we expressed and purified the ECD of short RPTPβ using a baculovirus expression system. The secreted short-RPTPβ ECD protein exhibited heterogenous glycosylation, running higher than its calculated MW of 85 kDa on SDS-PAGE (Fig. 4A, lane 1). The chondroitin sulfate proteoglycan mix, composed largely of phosphacan, was used as a control for these studies. This material ran as a high-molecular-weight protein mix above 300 kDa on SDS-PAGE (Fig. 4A, lane 2). Purified short-RPTPβ ECD was compared to phosphacan for its ability to act as a repulsive substrate in cell adhesion. U373 cells did not adhere effectively to tissue culture plates that were coated with short-RPTPβ ECD and adhered even less to plates that were coated with phosphacan. In both cases, the cells rounded up and displayed abnormal cell morphology. In contrast, negative control BSA did not inhibit cell attachment (Fig. 4B). Short-RPTPβ ECD behaved similarly, but was less effective than phosphacan. Pretreatment of U373 cells with short-RPTPβ ECD, phosphacan mix, or BSA, on the other hand, did not significantly affect cell adhesion (data not shown). Cells plated onto short-RPTPβ ECD or phosphacan mix rounded up and displayed abnormal cell morphology. The differences in cell morphology were visualized by staining with Alexa 594 phalloidin (Molecular Probes; Fig. 4C). Phalloidin specifically stains F-actin and thereby provides visual evidence of the disorganized cytoskeleton and cell rounding due to the short-RPTPβ ECD or phosphacan mix coating. These results indicate that the ECD of short RPTPβ retains much of the same functional capacity as phosphacan.
Fig. 4.
Short-RPTPβ ECD acts as a repulsive substrate for glioblastoma attachment. (A) Short-RPTPβ ECD was expressed in a baculovirus system and purified. Coomassie staining of short-RPTPβ ECD (10 μg) and phosphacan mix (10 μg) indicates the relative purity and size of these proteins. Short-RPTPβ ECD runs at about 85 kDa, and the phosphacan mix runs as a large-molecular-weight complex at >300 kDa. (B) U373 cells were plated and allowed to attach for 30 min onto plates coated with BSA, phosphacan, and short-RPTPβ ECD. Following three washes with PBS, the number of viable adherent cells was measured. Short RPTPβ-ECD inhibits binding of U373 cells slightly less effectively than phosphacan (error bars, ±SD; ** P < 0.005; *P < 0.05). (C) U373 cells were plated and allowed to attach for 30 min onto plates coated with BSA, phosphacan, and short-RPTPβ ECD. The cells were then stained with Alexa 594 phalloidin (Molecular Probes) to detect F-actin and cytoskeletal organization. Representative images were taken with a fluorescent microscope.
Signaling by Long and Short RPTPb
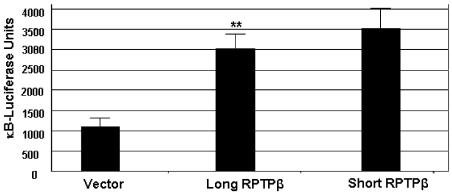
Overexpression of receptors such as RPTPβ can lead to forced dimerization that triggers signaling. In order to determine if forced overexpression of either long or short RPTPβ could trigger signaling events, a reporter assay was undertaken. We studied the transcription factor NF-κB because of its suggested role in integrating signals following cytoskeletal changes such as those involved in adhesion and migration. For similar reasons, β-catenin activation by RPTPβ/PTN has been studied (Meng et al., 2000). We also found that RPTPβ/PTN could activate β-catenin (data not shown). HEK 293 cells were transiently cotransfected with either vector, long RPTPβ or short RPTPβ, along with an NF-κB luciferase reporter construct and a β-galactosidase transfection control. The results from the reporter assay demonstrated that transfections with either long or short RPTPβ, but not pcDNA3.1 vector alone, activated the NF-κB response elements (Fig. 5). The NF-κB pathway has been implicated in a number of cell functions, including adhesion and migration of tumor cells. The difference in effect between long and short RPTPβ was insignificant, which suggests that both long and short RPTPβ have overlapping signaling capacities.
Fig. 5.
Long and short RPTPβ both activate NF-κB responsive elements to similar levels. HEK 293 cells were transiently cotransfected with vector or with long-RPTPβ or short-RPTPβ cDNA constructs, along with the NF-κB luciferase reporter construct and a β-galactosidase transfection control. Twenty-four hours posttransfection, the cells were lysed and assayed for luciferase and β-galactosidase activity by using the Dual-Lite Reporter Kit (Applied Biosystems). Luciferase activity was normalized to β-galactosidase activity (error bars, ±SD; ** P < 0.005).
Discussion
The differences between long and short RPTPβ have implications for the treatment of glioblastoma. We and others have demonstrated an upregulation of RPTPβ in glioblastoma (Mueller et al., 2003; Ulbricht et al., 2003). RPTPβ expression was correlated with increased malignancy (Ulbricht et al., 2003). In this study we demonstrate significant overlap in the expression patterns of long and short RPTPβ. The development of antibodies that specifically bind to the unique junction formed in short RPTPβ allows us to measure the relative distributions of both long and short RPTPβ in gliomas and normal brain tissue and to distinguish between the two forms. Phosphacan, while found in glioblastoma, is also present during brain development and following CNS injury. In contrast, both long and short forms of RPTPβ are predominantly expressed in glioblastoma and have very low and regional expression in normal brain. One strategy for the treatment of glioblastoma is the addition of therapeutic antibodies to the tumor resection site by convection-enhanced delivery. Immunoconjugates directed to unique regions of RPTPβ could specifically target tumor cells and destroy them. In addition, interfering with RPTPβ-mediated tumor cell migration, adhesion, and signaling could also contribute to the demise of tumor cells. Therefore, understanding the biology of long and short RPTPβ, as well as phosphacan, is important for the treatment of glioblastoma and potentially other cancers.
Differences in expression, structure, and functional effects between long and short RPTPβ can lead to qualitative and quantitative changes in signaling and biology. Despite overlapping expression patterns and similar signaling capacities, subtle differences may provide additional levels of biological complexity. Because the long and short RPTPβ forms described here share identical intracellular domains, they have the same capacity to signal. However, differences in their extracellular domains can also affect the biology of these proteins. In this study we demonstrate that short RPTPβ-ECD acts as a repulsive substrate, similar to phosphacan. This suggests that the primary cell adhesion–related regions reside in an area outside the 860 residue glycine-serine-rich domain encoded by exon 12. This glycine-serine-rich region, which is present in long RPTPβ and phosphacan, could have a role in modifying the binding of the CAH and FNIII domains with a host of RPTPβ ligands. In support of this notion, short RPTPβ does not contain chondroitin sulfate moieties (Nishiwaki et al., 1998). Further, chondroitinase treatment of phosphacan has been shown to decrease its effect on cell attachment by 25% (Milev et al., 1994). Given these observations, one can predict that short RPTPβ would be less effective at mediating adhesion than long RPTPβ, a fact borne out by our cell-based studies. Although short and long RPTPβ share the primary functional domains (CAH, FNIII, and phosphatase domains), the increased ability of long-RPTPβ-expressing cells to adhere to their ligands is apparent.
The differences in the long- and short-RPTPβ ECDs appear to translate into distinctive biological outcomes. This could be due to slight differences in the duration or intensity of the ligand-receptor-mediated signal or in the attributes of effector proteins. These subtle differences may collectively steer a cell toward migration or adhesion as opposed to proliferation or other outcomes. The work presented here also indicates that overexpression of both long and short RPTPβ leads to NF-κB transcriptional activity. Dysregulation of the NF-κB pathway has been linked to cancer. We suggest that the Akt kinase, a signal transducer common to both the NF-κB and β-catenin pathways, could provide key regulatory control over RPTPβ signaling. Both NF-κB and β-catenin have been implicated in cell adhesion and migration (among other things), and each could provide signals to control tumor biology. Any differences resulting from NF-κB and β-catenin activation by long and short RPTPβ have yet to be determined. In support of the notion that there may be a difference between the signals generated by long and short RPTPβ, PTN interacts with both long and short RPTPβ at high- and low-affinity sites (Maeda et al., 1996). Removal of the chondroitin sulfate groups from long RPTPβ (not found on short RPTPβ) reduced the affinity of PTN binding without changing the number of binding sites (Maeda et al., 1996), implying that PTN likely binds to short RPTPβ with lower affinity. The differences in binding would almost certainly affect signal intensity and duration. These modest differences can result in substantially altered outcomes. For instance, constitutive expression of PTN transforms NIH3T3 cells with a significant loss of contact inhibition, accompanied by decreased adhesion (Chauhan et al., 1993). This is particularly relevant because PTN is overexpressed in glioblastoma (Mueller et al., 2003). Conditions that cause differentiation or inhibit proliferation of rat C6 glioblastoma cells also cause downregulation of short RPTPβ relative to long RPTPβ and phosphacan (Canoll et al., 1996), which suggests that alterations in the expression level of long-and short-RPTPβ cells may affect migration, adhesion, proliferation, or other functions. Additional experiments are necessary to fully elucidate the subtle differences that may exist between the degrees of signaling pathway activation by long RPTPβ and short RPTPβ. These studies may also contribute to more effective therapeutic strategies for the treatment of glioblastoma and other cancers that express RPTPβ.
Footnotes
Abbreviations used are as follows: BSA, bovine serum albumin; CAH, carbonic anhydrase; ECD, extracellular domain; FNIII, fibronectin III domain; HRP, horseradish peroxidase; PBS, phosphate-buffered saline; RPTP, receptor-like PTP; PTN, pleiotrophin; PTP, protein tyrosine phosphatase; RPTPb, receptor protein tyrosine phosphatase beta; SDS, sodium dodecyl sulfate.
References
- Adamsky K, Schilling J, Garwood J, Faissner A, Peles E. Glial tumor cell adhesion is mediated by binding of the FNIII domain of receptor protein tyrosine phosphatase beta (RPTPβ) to tenascin C. Oncogene. 2001;20:609–618. doi: 10.1038/sj.onc.1204119. [DOI] [PubMed] [Google Scholar]
- Barnea G, Grumet M, Milev P, Silvennoinen O, Levy JB, Sap J, Schlessinger J. Receptor tyrosine phosphatase β is expressed in the form of proteoglycan and binds to the extracellular matrix protein tenascin. J Biol Chem. 1994;269:14349–14352. [PubMed] [Google Scholar]
- Canoll PD, Petanceska S, Schlessinger J, Musacchio JM. Three forms of RPTPβ are differentially expressed during gliogenesis in the developing rat brain and during glial cell differentiation in culture. J Neurosci Res. 1996;44:199–215. doi: 10.1002/(SICI)1097-4547(19960501)44:3<199::AID-JNR1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Chauhan AK, Li YS, Deuel TF. Pleiotrophin transforms NIH3T3 cells and induces tumors in nude mice. Proc Natl Acad Sci USA. 1993;90:679–682. doi: 10.1073/pnas.90.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood J, Heck N, Reichard F, Fraissner A. Phosphacan short isoform, a novel non-proteoglycan variant of phosphacan/receptor tyrosine phosphatase-beta, interacts with neuronal receptors and promotes neurite outgrowth. J Biol Chem. 2003;278:24164–24173. doi: 10.1074/jbc.M211721200. [DOI] [PubMed] [Google Scholar]
- Grumet M, Milev P, Sakurai T, Karthikeyan L, Bourdon MA, Margolis RK, Margolis RU. Interactions with tenascin and differential effects on cell adhesion of neurocan and phosphacan, two major chondroitin sulfate proteoglycans of nervous tissue. J Biol Chem. 1994;269:12142–12146. [PubMed] [Google Scholar]
- Levy JB, Canoll PD, Silvennoinen O, Barnea G, Morse B, Honegger AM, Huang JT, Cannizzaro LA, Park SH, Druck T. The cloning of a receptor-type protein tyrosine phosphatase expressed in the central nervous system. J Biol Chem. 1993;268:10573–10581. [PubMed] [Google Scholar]
- Maeda N, Nishiwaki T, Shintani T, Hamanaka H, Noda M. 6B4 proteoglycan/phosphacan, an extracellular variant of receptor-like protein-tyrosine phosphatase zeta/RPTPbeta, binds pleiotrophin/heparin-binding growth-associated molecule (HB-GAM) J Biol Chem. 1996;271:21446–21452. doi: 10.1074/jbc.271.35.21446. [DOI] [PubMed] [Google Scholar]
- Meng K, Rodríguez-Peña A, Dimitrov T, Chen W, Yamin M, Noda M, Deuel TF. Pleiotrophin signals increased tyrosine phosphorylation of β-catenin through inactivation of the intrinsic catalytic activity of the receptor-type protein tyrosine phosphatase β/ζ. Proc Natl Acad Sci USA. 2000;97:2603–2608. doi: 10.1073/pnas.020487997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev P, Friedlander DR, Sakurai T, Karthikeyan L, Flad M, Margolis RK, Grumet M, Margolis RU. Interactions of the chondroitin sulfate proteoglycan phosphacan, the extracellular domain of a receptor-type protein tyrosine phosphatase, with neurons, glia, and neural cell adhesion molecules. J Cell Biol. 1994;127:1703–1715. doi: 10.1083/jcb.127.6.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Kunkel P, Lamszus K, Ulbricht U, Lorente GA, Nelson AM, von Schack D, Chin DJ, Lohr SC, Westphal M, Melcher T. A role for receptor tyrosine phosphatase ζ in glioma cell migration. Oncogene. 2003;22:6661–6668. doi: 10.1038/sj.onc.1206763. [DOI] [PubMed] [Google Scholar]
- Mueller S, Lamszus K, Nikolich K, Westphal M. Receptor protein tyrosine phosphatase ζ as a therapeutic target for glioblastoma therapy. Expert Opin Ther Target. 2004;8:211–220. doi: 10.1517/14728222.8.3.211. [DOI] [PubMed] [Google Scholar]
- Nishiwaki T, Maeda N, Noda M. Characterization and developmental regulation of proteoglycan-type protein tyrosine phosphatase zeta/RPTPbeta isoforms. J Biochem. 1998;123:458–467. doi: 10.1093/oxfordjournals.jbchem.a021959. [DOI] [PubMed] [Google Scholar]
- Peles E, Schlessinger J, Grumet M. Multi-ligand interactions with receptor-like protein tyrosine phosphatase beta: Implications for intercellular signaling. Trends Biochem Sci. 1998;23:121–124. doi: 10.1016/s0968-0004(98)01195-5. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Friedlander DR, Grumet M. Expression of polypeptide variants of receptor-type protein tyrosine phosphatase beta: The secreted form, phosphacan, increases dramatically during embryonic development and modulates glial cell behavior in vitro. J Neurosci Res. 1996;43:694–706. doi: 10.1002/(SICI)1097-4547(19960315)43:6<694::AID-JNR6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Maeda N, Noda M, Marunouchi T. A chondroitin sulfate proteoglycan PTPzeta/RPTPbeta regulates the morphogenesis of Purkinje cell dendrites in the developing cerebellum. J Neurosci. 2003;23:2804–2814. doi: 10.1523/JNEUROSCI.23-07-02804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbricht U, Brockmann MA, Aiger A, Echerich C, Mueller S, Fillbrandt R, Westphal M, Lamszus K. Expression and function of the receptor protein tyrosine phosphatase zeta and its ligand pleiotrophin in human astrocytomas. J Neuropathol Exp Neurol. 2003;62:1265–1275. doi: 10.1093/jnen/62.12.1265. [DOI] [PubMed] [Google Scholar]