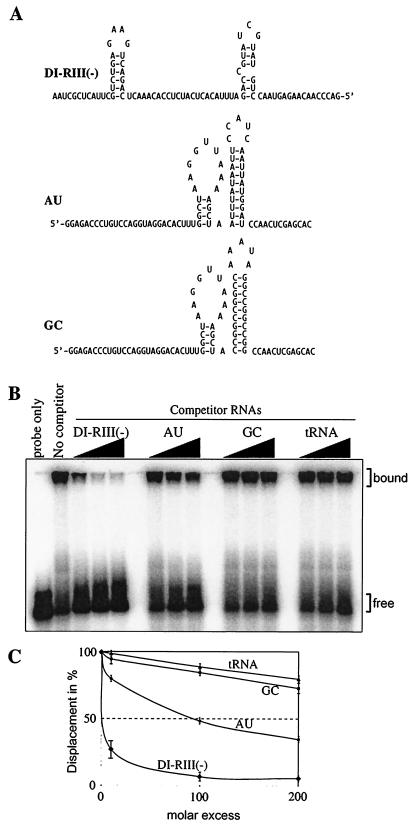
FIG. 4.
Testing binding preference of the recombinant p33 to RNA. (A) Predicted secondary structures of the competitor RNAs, including the 82-nt minus-stranded region III of TBSV [DI-RIII(-)] and the similarly sized artificial AU and GC templates (10). Note that the double- versus single-stranded regions are comparable in length in these competitor RNAs. (B) Unlabeled competitor RNAs at increasing amounts (in 10-, 100-, or 200-fold excess) were added to the mixture containing the labeled probe (82-nt minus-stranded region III; Fig. 1B) and 1 μM purified recombinant p33 and the bound complexes were analyzed in gel mobility shift assay. The tRNA was from yeast. (C) Graphical representation of data obtained in panel B. The quantification of the experiment was done as described in the legend to Fig. 3. The experiments were repeated twice.