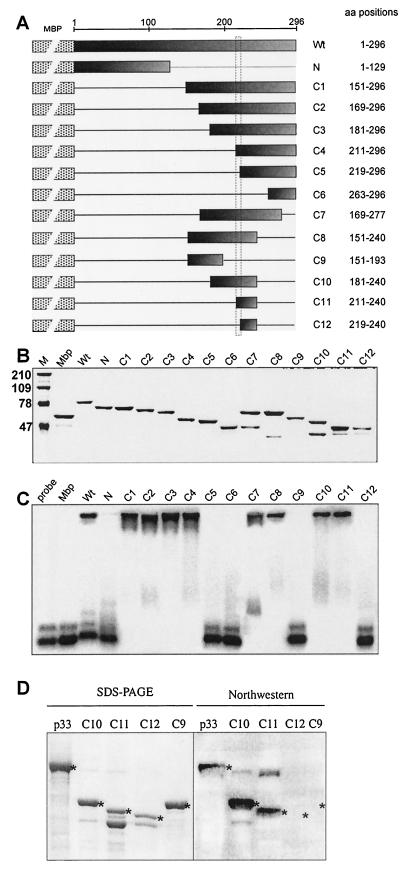
FIG.7.
Mapping the RNA-binding domain in the recombinant p33. (A) A schematic representation of the deletion derivatives of p33. The names of the constructs and the positions of the amino acids present in the truncated proteins are shown on the right. These truncated p33 proteins were expressed in E. coli as fusions to MBP (indicated schematically by a dotted box). The shaded boxes indicate the portions of p33 protein that were present in given expression constructs. The horizontal lines represent the deletions. (B) SDS-PAGE analysis of the purified recombinant proteins in a 10% polyacrylamide gel stained with Coomassie brilliant blue. The lane MW refers to molecular mass markers (in kilodaltons). (C) RNA binding activities of the truncated p33 proteins. The labeled RNA probe and the gel mobility shift assay were as described in the legend to Fig. 1B. Equimolar concentrations (2 μM) of proteins were used for the gel shift assay. (D) Northwestern analysis of selected truncated p33 proteins. The purified recombinant proteins (∼2 μg) were run in SDS-10% PAGE as shown in the left panel, transferred to a PVDF membrane, and then probed with a 32P-labeled probe [region III(-); Fig. 1B]. The positions in the Northwestern blot, which represent a particular recombinant protein, are marked with asterisks.