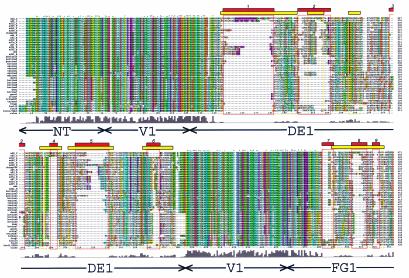
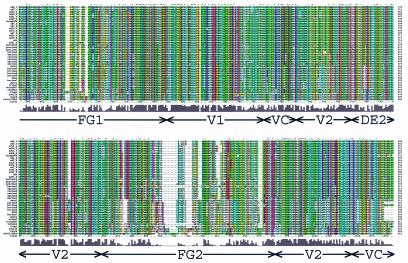
FIG. 2.
Hexon sequence alignment. Multiple-sequence alignment of 40 hexon sequences calculated with Clustal_X (64, 65). An alignment profile based on the structural alignment of two hexon crystal structures and three homology models was used to guide the multiple-sequence alignment. The default Clustal_X color scheme indicates significant features. All G (orange) and P (yellow) are colored. Frequent occurrences of a property at a sequence position are colored: hydrophobic, blue; hydrophobic tendency, light blue; basic, red; acidic, purple; hydrophilic, green; unconserved, white. Conserved positions are marked as complete (*), strong (:), or weak (.). A histogram indicates the conservation level, and the eight molecular regions are labeled. Previously assigned (15) HVRs (HVR1 to -9) are boxed in yellow, and the new assignments (HVR1 to -9) are boxed in red. A Clustal_X alignment file is available for download at http://bioinfo.wistar.upenn.edu/pub/RKB_hexon.aln.