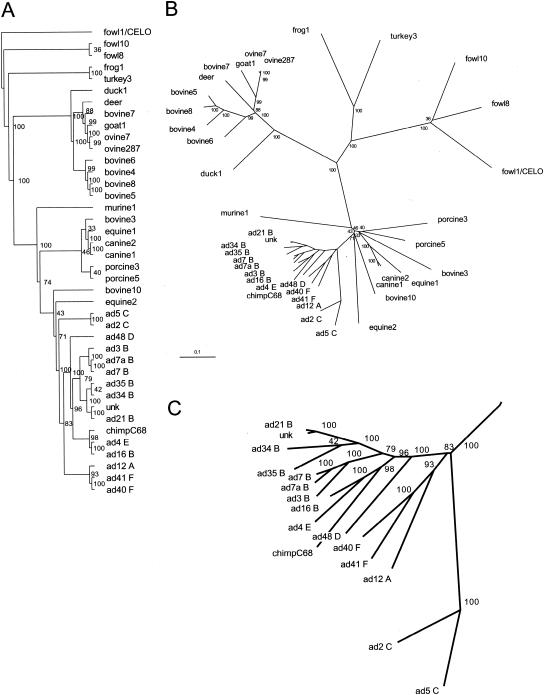
FIG. 4.
Phylogenetic analysis of adenovirus hexon. Phylogenetic trees of 40 full-length hexon sequences calculated with the Phylip program package (23). (A) An unrooted tree generated by parsimony analysis with PROTPARS and CONSENSE. (B) An unrooted tree generated by distance matrix analysis with PROTDIST (Dayhoff's PAM 001 scoring matrix), FITCH (global rearrangements option), and CONSENSE. Branch lengths are proportional to the number of substitutions, and the scale bar represents 10 mutations per 100 sequence positions. Bootstrap values for the 100 trials are indicated for each branch. (C) A close-up view of the Hominidae adenovirus hexon branches.