Abstract
A small open reading frame (ORF) in maedi-visna virus (MVV) and caprine arthritis encephalitis virus (CAEV) was initially named “tat” by analogy with a similarly placed ORF in the primate lentiviruses. The encoded “Tat” protein was ascribed the function of up regulation of the viral transcription from the long terminal repeat (LTR) promoter, but we have recently reported that MVV and CAEV Tat proteins lack trans-activation function activity under physiological conditions (S. Villet, C. Faure, B. Bouzar, G. Verdien, Y. Chebloune, and C. Legras, Virology 307:317-327, 2003). In the present work, we show that MVV Tat localizes to the nucleus of transfected cells, probably through the action of a nuclear localization signal in its C-terminal portion. We also show that, unlike the human immunodeficiency virus (HIV) Tat protein, MVV Tat was not secreted into the medium by transfected human or caprine cells in the absence of cell lysis but that, like the primate accessory protein Vpr, MVV and CAEV Tat proteins were incorporated into viral particles. In addition, analysis of the primary protein structures showed that small-ruminant lentivirus (SRLV) Tat proteins are more similar to the HIV type 1 (HIV-1) Vpr protein than to HIV-1 Tat. We also demonstrate a functional similarity between the SRLV Tat proteins and the HIV-1 Vpr product in the induction of a specific G2 arrest of the cell cycle in MVV Tat-transfected cells, which increases the G2/G1 ratio 2.8-fold. Together, these data strongly suggest that the tat ORF in the SRLV genomes does not code for a regulatory transactivator of the LTR but, rather, for a Vpr-like accessory protein.
Lentiviruses comprise a genus of complex, nononcogenic retroviruses that infect diverse mammalian hosts, including primates (human immunodeficiency virus [HIV] and simian immunodeficiency virus), carnivores (feline immunodeficiency virus), and ungulates (equine infectious anemia virus, bovine immunodeficiency virus, and the small-ruminant lentiviruses [SRLV]). In addition to the three essential retroviral genes, gag, pol, and env, lentiviruses carry a variable number of regulatory and accessory genes, most of which are situated between the end of pol and the beginning of env. The most complex of these viruses, HIV type 1 (HIV-1), encodes three regulatory genes, tat, rev, and vif, and three accessory genes, vpu, vpr, and nef. The three conserved open reading frames found between pol and env in the SRLVs maedi-visna virus (MVV) and caprine arthritis-encephalitis virus (CAEV) have classically been assigned to the regulatory genes tat, rev, and vif.
Some MVV-infected sheep progress to a stage of chronic debilitating disease affecting mainly the lungs (progressive interstitial pneumonia) and the central nervous system (progressive demyelinating encephalomyelitis) many years after infection (24, 25). Goats infected by the related CAEV can develop leukoencephalomyelitis (young kids) or chronic arthritis and mastitis (adult animals) (5, 6). In vivo, MVV and CAEV chiefly replicate in cells of the monocyte/macrophage lineage, and viral replication has been found to be dependent on the differentiation of monocytes into macrophages (10, 11). Unlike the primate and feline lentiviruses, MVV and CAEV do not cause immune deficiency in their infected hosts. This property correlates with their inability to cause productive infection of CD4+ T lymphocytes.
Some studies have reported an increase in the expression (15) of genes under the control of the MVV or CAEV long terminal repeat (LTR) or a stabilization of the corresponding mRNA (9) in the presence of the corresponding Tat peptide, but the increases in the levels of activity were inconstant and often low, ranging from 1.6-fold (12) to 46-fold (9). We have recently compared the transactivation activities of HIV-1 Tat with Tat proteins from MVV and CAEV on their homologous LTRs in different cell types, including macrophages, from natural host species and found little or no transactivation by SRLV Tat proteins under conditions in which the HIV-1 Tat produced a 60-fold increase in expression of the reporter gene (29). We concluded that transactivation of the homologous LTR was unlikely to be the principal function of the SRLV Tat proteins.
In the present study, we attempted to identify the possible natural activities of the SRLV Tat proteins through an analysis of their cellular localization and potential function. In transfected cells, the SRLV Tat protein is strongly concentrated in the nucleus, and this localization depends on the presence of signals on the C-terminal portion of the protein. In contrast to HIV-1 Tat, the SRLV Tat protein is never found in the culture supernatant of intact transfected human or ovine cells. It is, however, incorporated into both MVV and CAEV virus particles released from productively infected cells. Since all these characteristics are shared by the Vpr protein of HIV-1, we tested the SRLV Tat proteins for their ability to mediate an important function of HIV-1 Vpr, namely, cell cycle arrest in the G2 phase (22). We found that the ratio of cells in G2 to those in G1 increased 2.8-fold in unsynchronized cultures transfected with tat from MVV compared to that in mock-transfected controls. Taken together, the results presented in this paper clearly show that the lentiviruses of sheep and goats do not encode a potent transactivator of their own LTRs but, rather, a protein with functions closely related to those of the Vpr proteins of the primate lentiviruses.
MATERIALS AND METHODS
Plasmid constructs.
Plasmids directing the expression of green fluorescent protein (GFP) linked to the MVV Tat peptide or to the 5′ or 3′ portions of the molecule were constructed by inserting the relevant genetic elements between the XhoI and HindIII sites of pEGFP (Clontech, Hampshire, United Kingdom). The complete tat sequence was amplified from the p8Xsp5-RK1 (KV-1772) recombinant genomic plasmid (2) with primers UC1-tot and LC1-tot, and the truncated sequences were generated by using LC1-5′ in place of LC1-tot or UC1-3′ in place of UC1-tot.
A FLAG epitope-labeled Tat protein from HIV-1 and equivalent fusion proteins from MVV and CAEV were expressed from plasmids constructed by first inserting a 633-bp BglII-PstI fragment containing the cytomegalovirus promoter from pUT535 (Cayla, Montpelier, France) into plasmid BSK+ (Stratagene, Amsterdam, The Netherlands) and then adding a 158-bp SalI-ApaI fragment containing the simian virus 40 (SV40) polyadenylation signal from pUT614 (Cayla) to create the BSK-CMV-pA vehicle. The HIV-1 Tat sequence and the SRLV equivalents were tagged with the FLAG epitope by amplification from pSG5 Tat (kindly supplied by L. Gazzolo) for HIV-1, from p8Xsp5-RK1 for MVV, or from pK9 Kb for CAEV (19, 28) with 5′ primers containing the FLAG coding sequence (H-Flag5′, V-Flag5′, and C-Flag5′, respectively) and the relevant 3′ primers (Tat HIV3, LC1tot, and Tat 04, respectively) as follows: UC1-tot, 5′CCCTCGAGGCGAAGAAGTACCAAGAAG3′; LC1-tot, 5′CCAAGCTTATCACAGTTCAACATT TC3′; LC1-5′, 5′CCAAGCTTTCATTCGCCAGGTACTCTT 3′; UC1-3′, 5′CCCTCGAGGCAGACTACAACGATGGCT 3′; C-Flag5′, 5′GGAATTCATGGACTACAAGGACGACGATGACAGTGAAGAACTGCCTCAAAGAAGGGAGACACATCCAGAAGAACTTGTAAGGAACGTACGGGAAAGAGAAAG3′; Tat 04, 5′CCAAGCTTGATTATGTTCCCCACCC3′; V-Flag5′, 5′GGAATTCATGGACTACAAGGACGACGATGACAAGGAAGAAGTACAAGAAGAAGCCAGGAGGCTTAGTAGAAGTAGAGGGAGTATTTCAATTTTATGAAGA3′; H-Flag5′, 5′TGAATTCATGGACTACAAGGACGACGATGACAAGGAGCCAGTAGATCCTAGCTAGAGCCCTGGAAGCATCCAGGAAGT CAGCCTAAAACTGCTTGTACCACTT3′; and Tat HIV3, 5′CCAAGCTTCGAGCTATTCCTTC3′.
Cells.
Human epithelioid carcinoma HeLa cells were maintained at 37°C in 5% CO2 in Dulbecco's minimal essential medium (Life Technologies, Cergy Pontoise, France) supplemented with 10% fetal calf serum (Life Technologies). Large T immortalized goat embryo fibroblasts (TIGEF) (7) and goat synovial membrane (GSM) cells were maintained in Eagle's minimal essential medium (Life Technologies) supplemented with 10% fetal calf serum.
Transfection of plasmid DNAs and virus production.
HeLa cells or TIGEF were seeded into 60-mm-diameter (2 · 105 cells/plate) or 100-mm-diameter (5 · 105 cells/plate) tissue culture petri dishes (Elvetech, Lyon, France) and maintained in culture for 24 h before transfection with Effectene (Qiagen, Courtaboeuf, France), according to the manufacturer's recommendations. Briefly, for a 60-mm dish, 8 μl of enhancer was added to 1 μg of the relevant plasmid DNA in DNA condensation buffer and incubated for 3 min at room temperature prior to the addition of 10 μl of Effectene reagent. After a further 10 min at room temperature, the mixture was added to the culture dish and incubated for 5 h before the medium was changed. For 100-mm dishes, double quantities of DNA and reagents were used.
CAEV-pBSCA virus stock was generated by Lipofectamine (Life Technologies) transfection of 5 μg of plasmid pBSCA DNA into 5 · 105 GSM cells which had been seeded into a 25-cm2 tissue culture flask 24 h previously (7). After 18 h and three washes with phosphate-buffered saline (PBS), the medium was replaced and the virus-containing supernatant medium was collected at 3, 5, and 7 days posttransfection. Supernatants were stored at −80°C after they were clarified through 45-μm-pore-size membranes (Millipore, St. Quentin, France). MVV strain K1514 was produced on GSM cells under standard conditions (7).
Immunoprecipitation assay.
Supernatant (1 ml) was collected from 60-mm culture dishes 48 h after transfection and immediately mixed with 100 μl of 10× radioimmunoprecipitation assay (RIPA) buffer (0.5 M Tris [pH 7.5], 0.5 M NaCl, 5% deoxycholic acid, 2% sodium dodecyl sulfate [SDS], 0.1 M EDTA). The cell layer was then scraped into 1 ml of 1× RIPA buffer, transferred into a 1.5-ml Eppendorf tube, and clarified by centrifugation for 10 min at 1,300 × g. Specific antigens were sought by adding 1 μl of anti-FLAG antibody (Bio-Rad, Marnes-la-Coquette, France) or 5 μl of anti-SV40 large T antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) in 20 μl of 1× RIPA buffer to 50 μl of a 6% suspension of protein A-Sepharose (Sigma, La Verpillère, France); this mixture was then added to the cell supernatant or lysate preparations in a fresh Eppendorf tube, which was stored overnight at 4°C on a rocker. The Sepharose beads were then recovered by centrifugation at 500 × g for 2 min and washed five times in 1× RIPA buffer; the captured antigens were released for immunoblot analysis by resuspension in 50 μl of 2× reducing buffer (80 mM Tris [pH 6.8], 12% glycerol, 2% SDS, 2% β-mercaptoethanol, bromophenol blue).
Preparation of virions and cell lysates.
To test whether tagged SRLV Tat proteins could be incorporated into virions, TIGEF in 100-mm dishes were first infected with either CAEV or MVV (multiplicity of infection [MOI], 1) for 2 h and then cultured in fresh medium for 24 h. The TIGEF were then transfected with the relevant plasmids encoding FLAG-Tat-tagged proteins as described above, and the culture supernatants were collected 72 h later. After clarification at 1,300 × g for 10 min, virions were pelleted from the supernatants by centrifugation at 100,000 × g for 45 min at 4°C and then recovered in 45 μl of lysis buffer (10 mM Tris [pH 7.5], 0.15 M NaCl, 2 mM EDTA, 0.5% Nonidet P-40). The cell layers were washed twice with PBS and disrupted with trypsin. The cells were recovered by centrifugation at 500 × g for 5 min, resuspended in 100 μl of cold lysis buffer, and maintained on ice for 4 min before clarification in a microcentrifuge for 5 min. The cell lysates and virion preparations were stored at −80°C until used.
For density gradient analysis, six culture dishes (diameter, 100 mm) of TIGEF were infected and then transfected as described above. At 72 h posttransfection, the supernatants were pooled and the virions were pelleted by centrifugation at 100,000 × g for 45 min. The pellets were resuspended in 500 μl of 10 mM Tris (pH 7.5)-100 mM NaCl-1 mM EDTA and layered onto an 11-ml linear sucrose gradient (20 to 60% [wt/vol] sucrose in 10 mM Tris [pH 7.5]-100 mM NaCl-1 mM EDTA). The gradient was centrifuged for 16 h at 100,000 × g at 10°C, and 12 fractions (0.9 ml) were collected from the top of the tube. Materials contained in each fraction were pelleted and dissolved in 30 μl of lysis buffer and then subjected to immunoblot analysis.
Immunoblot analysis.
Proteins from lysates (10 μl) or supernatants (20 μl) in sample reducing buffer were separated on 16.5% polyacrylamide gels in the presence of SDS. The proteins were transferred from the gel to nitrocellulose transfer membranes (Schleicher & Schuell, Ecquevilly, France), which were probed with monoclonal antibodies directed against the FLAG epitope (diluted 10,000 times; Bio-Rad), SV40 large T antigen (diluted 1,000 times; Santa Cruz Biotechnology), or CAEV capsid protein (CAEP8B1 monoclonal antibody, diluted 1,000 times; VMRD Inc., Pullman, Wash.) Bound antibodies were detected by incubation with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (diluted 3,000 times; Bio-Rad) and visualized by enhanced chemiluminescence (ECL kit; Amersham, Saclay, France) according to the manufacturer's instructions.
Flow cytometry analysis of G2 arrest of cell cycle.
The cell cycle was studied quantitatively by flow cytometric determination of the nuclear DNA content after cells were stained with propidium iodide (Sigma) as previously described (23). Briefly, 5 · 105 transfected cells were harvested following dissociation with trypsin; they were then rinsed and resuspended in 1 ml of PBS. Cell suspensions were then fixed by the addition of 500 μl of 1% paraformaldehyde and a 30-min incubation in ice. The fixed cells were rinsed once with PBS, incubated for 30 min at room temperature in a solution containing RNase A (10 mg/ml), and then stained by the addition of 2 μl of 10-mg/ml propidium iodide solution. The DNA content was finally analyzed for more than 10,000 events by FACScan flow cytometry (Becton Dickinson, Le Pont de Claix, France). The cell cycle distribution of fluorescent transfected cells was analyzed with Cell Quest software. Background levels of fluorescence were determined by using mock-transfected cells.
RESULTS
Subcellular localization of MVV Tat protein.
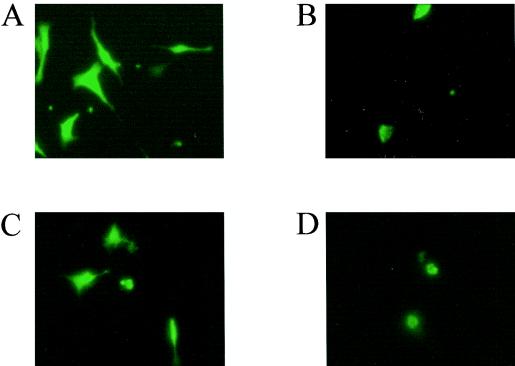
We evaluated the intracellular localization of an SRLV Tat protein by comparing the distribution of fluorescence in TIGEF transfected with a plasmid (pGFP-Tat) expressing a GFP-MVV Tat fusion protein with that in cells transfected by the pGFP plasmid, which expresses only the fluorescent marker. GFP was distributed throughout the cytoplasm and nucleus of the transfected cells, as observed by fluorescence microscopy at 48 h posttransfection (Fig. 1A), whereas the GFP-Tat fusion protein was essentially present in the nucleus (Fig. 1B). To establish which portion of the MVV Tat protein was responsible for this localization, we constructed two further plasmids expressing either the N-terminal (activator) portion or the leucine-rich C-terminal portion of the protein as fusions with GFP. TIGEF transfected with the construct expressing the N-terminal portion of MVV Tat and examined 48 h later showed a distribution of fluorescence throughout the whole cell (Fig. 1C), like the distribution for GFP alone, whereas the cells transfected with the construct expressing the C-terminal portion of MVV Tat showed a nuclear localization of fluorescence (Fig. 1D), like the distribution for the intact Tat protein.
FIG. 1.
Localization of wild-type and mutant MVV Tat-GFP fusion proteins. The TIGEF cell line was transfected with pGFP (A), pGFP-Tat (B), pGFP-5′ (C), and pGFP-3′ (D) plasmid constructs; 48 h posttransfection, the cells were observed by fluorescence microscopy. Magnification, ×400.
MVV Tat protein is not released from transfected cells.
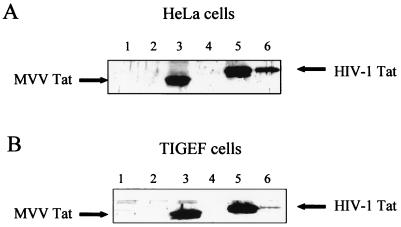
It has been clearly demonstrated that the HIV-1 Tat protein is secreted into the culture medium by infected, or transfected, cells (8), so we investigated whether an SRLV Tat protein was also present in the supernatant medium of our transfected cells. To facilitate detection of the Tat proteins, they were expressed as fusion proteins with the FLAG epitope by the plasmids pTat-Flag-MVV and pTat-Flag-HIV. At 48 h after transfection of HeLa cells, the supernatant media were collected and analyzed by immunoblot analysis following specific immunoprecipitation with the antibody to the FLAG epitope. The cell monolayer was also lysed and subjected to the same analysis. As expected, the HIV-1 Tat protein was easily detectable in both the cell lysate and the supernatant medium, but MVV Tat was absent from the supernatant medium, although clearly present in the cell lysate (Fig. 2A). Because the secretion of the Tat protein into the supernatant might be a property that is characteristic of natural host cells for the virus, we repeated the experiment using caprine TIGEF as hosts for the two plasmids. Again, the HIV-1 Tat protein was clearly present in the supernatant, although in a smaller quantity than when it was expressed in HeLa cells, and the MVV Tat protein was seen only in the cell lysate (Fig. 2B). TIGEF constitutively express the SV40 large T antigen, which was used for their immortalization, so we probed the samples with the antibody to SV40 large T as an internal control. The antigen was clearly present in the cell lysates and absent from either supernatant, suggesting that the secretion of HIV-1 Tat into the medium was a specific event and not simply due to cellular leakage (data not shown). In summary, MVV Tat, unlike HIV-1 Tat, is not actively secreted into the culture medium from transfected human or caprine cell cultures.
FIG. 2.
Release of the Tat protein in the supernatant of Tat-expressing cells. The human HeLa (A) or goat TIGEF (B) cell lines were transfected with the pTat-Flag-MVV and pTat-Flag-HIV plasmid constructs. At 48 h posttransfection, proteins from supernatants (lanes 2, 4, and 6) and cell lysates (lanes 1, 3, and 5) were immunoprecipitated using an anti-FLAG monoclonal antibody. Immunoblots of immunoprecipitated fractions from mock-transfected (lanes 1 and 2), pTat-Flag-MVV-transfected (lanes 3 and 4), and pTat-Flag-HIV-transfected (lanes 5 and 6) cells were performed with a specific monoclonal antibody directed against the FLAG epitope.
Incorporation of tagged Tat into virions.
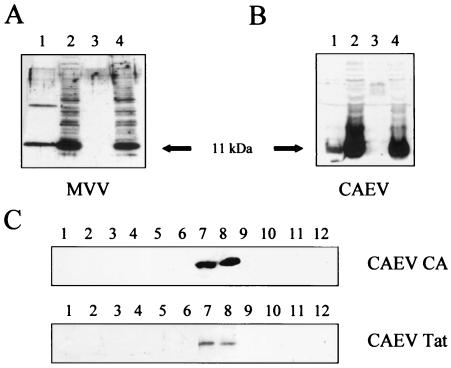
We next tested whether SRLV Tat proteins, although not released free into the supernatant of transfected cells, were incorporated in the virions on productive infection. TIGEF were infected at an MOI of 1 with either MVV or CAEV and then transfected 24 h later with pTat-Flag-MVV or the equivalent pTat-Flag-CO plasmids, respectively. Supernatants and cell lysates were collected 72 h later and analyzed by immunoblotting with the anti-FLAG monoclonal antibody. Control samples were mock infected and then transfected similarly. As shown in Fig. 3A and B, cell lysates from cells transfected with MVV or CAEV Tat-Flag plasmids contained an abundant 11-kDa band recognized by the anti-FLAG antibody, whether they were infected with a replicative virus or not. In contrast, an 11-kDa band was present only in the supernatants of infected cells and never in those of the noninfected controls.
FIG. 3.
MVV and CAEV Tat proteins are associated with viral particles. TIGEF were inoculated (MOI, 1) with MVV (A) or CAEV (B and C) or were mock infected. At 24 h postinoculation, the cells were transfected with pTat-Flag-MVV (A) and pTat-Flag-CO (B and C) plasmids. At 48 h posttransfection, the supernatants were collected and virions were harvested from pellets by ultracentrifugation; cell lysates were then obtained following treatment with the lysing solution. (A and B) Proteins from the supernatants (lanes 1 and 3) and cell lysates (lanes 2 and 4) were analyzed by immunoblot assay by using an antibody directed against the FLAG epitope (diluted 10,000 times). Lanes 1 and 2, cells infected with MVV (A) or CAEV (B); lanes 3 and 4, mock-infected cells. (C) Pellets of virions were resuspended and loaded on the top of a linear sucrose gradient. After 16 h of ultracentrifugation, 12 fractions were collected and proteins were extracted and then analyzed by immunoblot assay with an anti-p25 Gag antibody to reveal the major CAEV capsid protein (diluted 1,000 times) or an anti-FLAG antibody (diluted 10,000 times) to reveal the CAEV Tat protein.
Since the result described above suggested that SRLV Tat proteins might be incorporated into budding virions, we analyzed the supernatant from CAEV-infected and transfected cells by density gradient ultracentrifugation. Cells were infected and transfected as described above, the supernatant medium was subjected to high-speed centrifugation, and the resuspended pellet was placed on the top of a linear sucrose gradient as described in Materials and Methods. After ultracentrifugation, the gradient was divided into 12 fractions, and these were analyzed for the CAEV Tat fusion protein with the anti-FLAG antibody and for the major CAEV capsid antigen by using an anti-CAEV p25 Gag antibody. Proteins were separated by SDS-polyacrylamide gel electrophoresis and analyzed by immunoblotting with the antibodies mentioned above, and the Tat-FLAG fusions always colocalized with the CAEV capsid proteins (Fig. 3C). This strongly suggests that the SRLV Tat proteins are incorporated into the viral particle.
Expression of Tat increases the proportion of cells arrested in the G2 phase of the cell cycle.
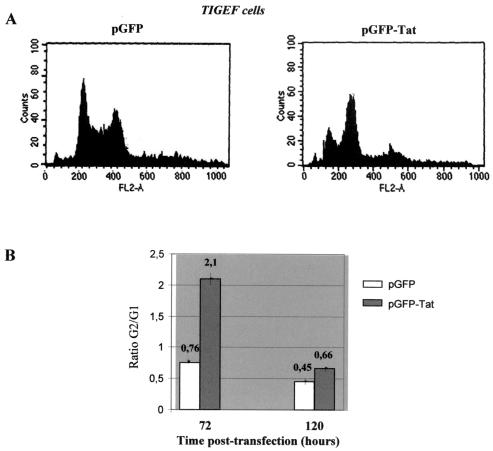
The possible effect of the SRLV Tat proteins on the cell cycle of the infected cells was investigated by using TIGEF transiently transfected with an MVV Tat-GFP fusion protein and then fixed and stained with propodium iodide. This procedure allowed simultaneous evaluation of the expression of the Tat fusion and of the quantity of cellular DNA. Control cells were transfected with GFP alone. DNA profiles were determined at 72 and 120 h posttransfection. At 72 h, for cells transfected with GFP alone, 46% of the transfected cells were in the G1/S phase and 35% were in the G2/M phase of their cycles; for Tat-transfected cells, only 24% were in the G1/S phase compared to 51% in G2/M at the same time (Fig. 4A). Assuming that the time spent in M and S is unchanged, this represents an increase of 2.8-fold in the ratio of cells in G2 to G1 in Tat-transfected cells. At 120 h posttransfection, the effect was less pronounced, but the Tat-transfected cells still showed a 1.5-fold increase in the G2/G1 ratio.
FIG. 4.
G2 arrest of cell cycle in Tat-expressing cells. TIGEF were transfected with plasmid pGFP as a negative control or with plasmid pGFP-Tat. Cells were harvested 72 and 120 h posttransfection, and the DNA was stained with propidium iodide. Cells were analyzed by flow cytometry of more than 10,000 events with Cell Quest analysis software. Histograms of the flow cytometry analysis of the DNA content in transfected TIGEF harvested 72 h posttransfection are shown in panel A. The ordinate indicates the number of cells, and the abscissa indicates the DNA content. (B) The ratios of the percentages of cells in G2/G1 phases of the cell cycle are shown. Values shown at the top of each bar represent the mean value of results of at least three independent transfection experiments.
DISCUSSION
It has recently been shown that the proteins translated from the open reading frames identified as tat in the SRLV do not function primarily as up regulators of transcription from the viral LTR, as do the Tat peptides of the primate lentiviruses (29). In the present study we pursued a comparison of the SRLV Tat proteins with the different primate lentivirus accessory and regulatory proteins. We used vectors expressing SRLV Tat proteins fused to identifiable markers to examine the intracellular distribution of the protein, its secretion into the supernatant medium, and its possible incorporation into released virions. Finally, we evaluated the capacity of an SRLV Tat peptide to mediate an important function of an HIV-1 accessory protein which is the specific arrest in the G2 phase of the cell cycle.
Cells transfected with a plasmid encoding native MVV Tat fused to GFP showed a clear nuclear fluorescence, with occasional, possibly perinuclear, patches (Fig. 1B). We did not observe the nucleolar concentration typical of HIV-1 Tat (26, 27). A similar pattern of expression is obtained when GFP is fused to the C-terminal, leucine- and cysteine-enriched portion of the MVV Tat protein, but the N-terminal portion (the so-called activator domain) does not direct a nuclear concentration. The C-terminal portion of MVV Tat begins within the leucine-rich conserved region and extends to the natural terminator following another well-conserved cysteine-rich domain. The intermediate region, although globally not well conserved between different viral isolates, always contains many basic amino acids, often in individually conserved positions, which may constitute a nuclear localization signal. Studies involving site-directed mutagenesis of these amino acids in the present construct may clarify their role.
Using the FLAG epitope fused to the SRLV Tat proteins, we showed that these proteins are not secreted into the medium from transfected cells, unlike tagged HIV-1 Tat, which is secreted from the same cells under the same conditions. In natural infections, HIV-1 Tat excreted from an infected cell is believed to enter neighboring cells (8) and to contribute to the development of pathology. The amino acid sequence of HIV-1 Tat contains no identifiable secretion signal (14), and such leaderless secreted proteins are believed to be externalized through the endoplasmic reticulum and Golgi apparatus by a nonclassical pathway which is still poorly understood (1). We show in this study that FLAG-tagged HIV-1 Tat is secreted from transfected caprine cells (TIGEF), although less efficiently than from human (HeLa) cells. This suggests that the mechanism for export of leaderless secreted proteins is conserved in mammals but that HIV-1 Tat is specifically better adapted to the human mechanism. The absence of MVV Tat from the supernatants of either human or caprine cells suggests that it does not employ this cellular mechanism.
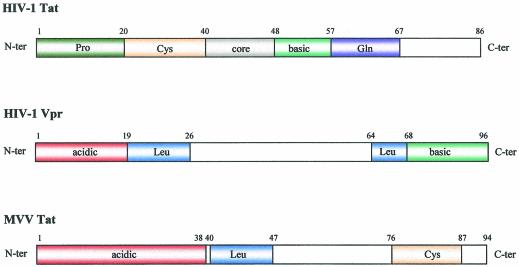
The findings described above indicate important functional differences between HIV-1 Tat and the SRLV Tat proteins, which are emphasized by the finding that the latter are exported from actively infected cells in association with the viral particles (Fig. 3). A comparison with other regulatory and accessory proteins of the primate lentiviruses showed a considerable similarity to the Vpr accessory protein. This is a small 96-amino-acid (14-kDa) protein which is expressed in the nucleus and is exported in the viral particles budding from infected cells (4, 17). FLAG labeling of HIV-1 Vpr did not alter its distribution in infected cells or viral particles (19). Characteristic functions of HIV-1 Vpr include arrest of infected cells in the G2/M phase of their cycle (3, 16) by inhibition of p34cdc2 activity (13, 22), participation in nuclear import of the viral preintegration complex in nondividing cells (20, 21), and a weak (two- to sevenfold) trans-activation of the viral LTR (30). Alignment of the primary amino acid sequences of the SRLV Tat proteins and HIV-1 Vpr protein showed that both sequences contain an acidic N-terminal domain, a central leucine-rich domain, and an arginine-rich C-terminal portion (Fig. 5). The C-terminal portion of the SRLV Tat is usually described as cysteine rich; however, it can be seen that this sequence is also relatively rich in arginine residues. Thus, the primary structures of SRLV Tat and HIV-1 Vpr proteins are very similar, and the same main domains are conserved. In contrast, a corresponding examination of the primary structure of HIV-1 Tat protein revealed no noteworthy similarity with the structures of either SRLV Tat or HIV-1 Vpr.
FIG. 5.
Comparison of the primary structures of the SRLV and HIV-1 Vpr and Tat proteins. The putative domains of MVV Tat, HIV-1 Tat, and HIV-1 Vpr are shown as stained boxes. The HIV-1 Tat protein contains an N-terminal proline-rich domain (Pro), a cysteine-rich domain (Cys), a core, a basic domain (basic), and a glutamine-rich domain (Gln). The HIV-1 Vpr protein contains an N-terminal acidic domain (acidic), two leucine-rich domains (Leu), and a basic domain (basic). MVV Tat protein contains an N-terminal acidic domain (acidic), a leucine-rich domain, and a cysteine-rich domain.
This resemblance of the SRLV Tat proteins to HIV-1 Vpr is reinforced by our finding that cells transfected with a plasmid expressing an MVV Tat-GFP fusion protein accumulate in the G2/M phase of the cell cycle, with a 2.8-fold increase in cells in G2/M compared to those in G1/S at 72 h posttransfection. The effect persisted at lower levels until at least 120 h posttransfection. This result indicates that MVV Tat, like HIV-1 Vpr, induces specific arrest in the G2 phase of the cell cycle.
In conclusion, the different properties of the SRLV Tat proteins, including primary amino acid sequence, cellular and virion localization, absence of strong transactivation of the viral LTR, and cellular arrest in the G2 phase of the cycle, lead to the conclusion that this protein is better considered an accessory protein similar to the Vpr protein of the primate lentiviruses and that the SRLV do not encode a veritable Tat protein.
Acknowledgments
This work was supported by a research grant from the Centre National de la Recherche Scientifique, Institut National de la Recherche Agronomique. We thank the Ministère de la Recherche for the fellowship (S.V.).
We thank L. Gazzolo for kindly providing the pSG5 Tat plasmid. We also thank Timothy Greenland for helpful discussion and for writing.
REFERENCES
- 1.Andrei, C., C. Dazzi, L. Lotti, M. R. Torrisi, G. Chimini, and A. Rubartelli. 1999. The secretory route of the leaderless protein interleukin 1β involves exocytosis of endolysosome-related vesicles. Mol. Biol. Cell 10:1463-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andresson, O. S., J. E. Elser, G. J. Tobin, J. D. Greenwood, M. A. Gonda, G. Georgsson, V. Andresdottir, E. Benediktsdottir, H. M. Carlsdottir, and E. O. Mantyla. 1993. Nucleotide sequence and biological properties of a pathogenic proviral molecular clone of neurovirulent visna virus. Virology 193:89-105. [DOI] [PubMed] [Google Scholar]
- 3.Bartz, S. R., M. E. Rogel, and M. Emerman. 1996. Human immunodeficiency virus type 1 cell cycle control: Vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J. Virol. 70:2324-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, E. A., G. Dehni, J. G. Sodroski, and W. A. Haseltine. 1990. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J. Virol. 64:3097-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cork, L. C., W. J. Hadlow, T. B. Crawford, J. R. Gorham, and R. C. Piper. 1974. Infectious leukoencephalomyelitis of young goats J. Infect. Dis. 129:134-141. [DOI] [PubMed] [Google Scholar]
- 6.Crawford, T. B., D. S. Adams, W. P. Cheevers, and L. C. Cork. 1980. Chronic arthritis in goats caused by a retrovirus. Science 207:997-999. [DOI] [PubMed] [Google Scholar]
- 7.Da Silva Teixeira, M. F., V. Lambert, L. Mselli-Lakahl, A. Chettab, Y. Chebloune, and J. F. Mornex. 1997. Immortalization of caprine fibroblasts permissive for replication of small ruminant lentiviruses. Am. J. Vet. Res. 58:579-584. [PubMed] [Google Scholar]
- 8.Ensoli, B., L. Buonaguro, G. Barillari, V. Fiorelli, R. Gendelman, R. A. Morgan, P. Wingfield, and R. C. Gallo. 1993. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J. Virol. 67:277-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gdovin, S. L., and J. E. Clements. 1992. Molecular mechanisms of visna virus Tat: identification of the targets for transcriptional activation and evidence for a posttranscriptional effect. Virology 188:438-450. [DOI] [PubMed] [Google Scholar]
- 10.Gendelman, H. E., O. Narayan, S. Kennedy-Stoskopf, P. G. E. Kennedy, Z. Ghotbi, J. E. Clements, J. Stanley, and G. Pezeshkpour. 1986. Tropism of sheep lentiviruses for monocytes: susceptibility to infection and virus gene expression increase during maturation of monocytes to macrophages. J. Virol. 58:67-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gendelman, H. E., O. Narayan, S. Molineaux, J. E. Clements, and Z. Ghotbi. 1985. Slow, persistent replication of lentiviruses: role of tissue macrophages and macrophage precursors in bone marrow. Proc. Natl. Acad. Sci. USA 82:7086-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gourdou, I., K. Mabrouk, G. Harkiss, P. Marchot, N. Watt, F. Hery, and R. Vigne. 1990. Neurotoxicity in mice due to cysteine-rich parts of visna virus and HIV-1 Tat proteins. C. R. Acad. Sci. Ser. III 311:149-155. [PubMed] [Google Scholar]
- 13.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helland, D. E., J. L. Welles, A. Caputo, and W. A. Haseltine. 1991. Transcellular transactivation by the human immunodeficiency virus type 1 tat protein. J. Virol. 65:4547-4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hess, J. L., J. A. Small, and J. E. Clements. 1989. Sequences in the visna virus long terminal repeat that control transcriptional activity and respond to viral trans-activation: involvement of AP-1 sites in basal activity and trans-activation. J. Virol. 63:3001-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jowett, J. B. M., V. Planelles, B. Poon, N. P. Shah, M.-L. Chen, and I. S. Y. Chen. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol. 69:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, Y.-L., P. Spearman, and L. Ratner. 1993. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J. Virol. 67:6542-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mselli-Lakhal, L., C. Favier, K. Leung, F. Guiguen, D. Grezel, P. Miossec, J.-F. Mornex, O. Narayan, G. Quérat, and Y. Chebloune. 2000. Lack of functional receptors is the only barrier that prevents caprine arthritis-encephalitis virus from infecting human cells. J. Virol. 74:8343-8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paxton, W., R. I. Connor, and N. R. Landau. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J. Virol. 67:7229-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Popov, S., M. Rexach, L. Ratner, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates docking of the HIV-1 preintegration complex to the nuclear pore complex. J. Biol. Chem. 273:13347-13352. [DOI] [PubMed] [Google Scholar]
- 21.Popov, S., M. Rexach, G. Zybarth, N. Reiling, M. A. Lee, L. Ratner, C. M. Lane, M. S. Moore, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 17:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Re, F., D. Braaten, E. K. Franke, and J. Luban. 1995. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J. Virol. 69:6859-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogel, M. E., L. I. Wu, and M. Emerman. 1995. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J. Virol. 69:882-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigurdsson, B., and P. A. Palsson. 1958. Visna of sheep. A slow demyelinating infection. Br. J. Pathol. 39:519-528. [PMC free article] [PubMed] [Google Scholar]
- 25.Sigurdsson, B., P. A. Palsson, and H. Grimsson. 1957. Visna, a demyelinating transmissible disease of sheep. J. Neuropathol. Exp. Neurol. 16:389-403. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian, T., R. Govindarajan, and G. Chinnadurai. 1991. Heterologous basic domain substitutions in the HIV-1 Tat protein reveal an arginine-rich motif required for transactivation. EMBO J. 10:2311-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian, T., M. Kuppuswamy, L. Venkatesh, A. Srinivasan, and G. Chinnadurai. 1990. Functional substitution of the basic domain of the HIV-1 trans-activator, Tat, with the basic domain of the functionally heterologous Rev. Virology 176:178-183. [DOI] [PubMed] [Google Scholar]
- 28.Turelli, P., G. Pétursson, F. Guiguen, J.-F. Mornex, R. Vigne, and G. Queérat. 1996. Replication properties of dUTPase-deficient mutants of caprine and ovine lentiviruses. J. Virol. 70:1213-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villet, S., C. Faure, B. Bouzar, G. Verdier, Y. Chebloune, and C. Legras. 2003. Lack of trans-activation function for maedi visna virus (MVV) and caprine arthritis encephalitis virus (CAEV) Tat proteins. Virology 307:317-327. [DOI] [PubMed]
- 30.Wang, L., S. Mukherjee, F. Jia, O. Narayan, and L. J. Zhao. 1995. Interaction of virion protein Vpr of human immunodeficiency virus type 1 with cellular transcription factor Sp1 and trans-activation of viral long terminal repeat. J. Biol. Chem. 270:25564-25569. [DOI] [PubMed] [Google Scholar]