FIG. 1.
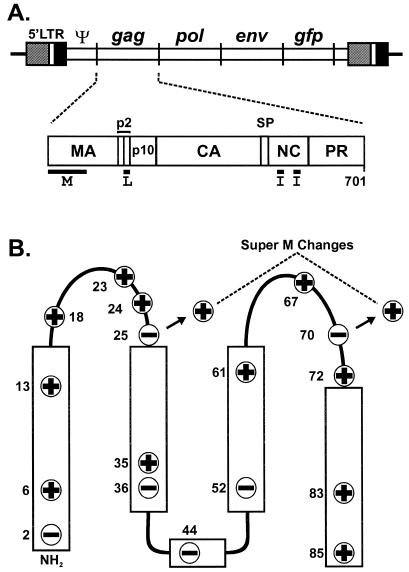
Location of the Super M substitutions in RSV Gag. (A) Diagram of the RSV proviral DNA used in these experiments (top) and the RSV Gag polyprotein (bottom). The 5′ long terminal repeat (LTR) promotes transcription of genome-length mRNA, which can be encapsidated or used for the synthesis of Gag and Gag-Pol proteins. In addition, full-length vRNA can be spliced for the synthesis of Env glycoproteins and, in the recombinant virus used here, for the synthesis of GFP. Intact Gag polyproteins drive budding and are subsequently cleaved by the viral PR into the mature products: MA, CA, NC, etc. The locations of the domains required for budding are indicated below Gag. The M domain is essential for plasma membrane targeting. The I domains promote Gag-Gag interactions and the assembly of dense particles. The L domain is required for a late step in budding. (B) The secondary structure of the RSV M domain consists of five helices (rectangles) connected by flexible loops. The locations of the 11 basic and 6 acidic residues in the M domain are depicted according to their charge. The two acidic-to-basic substitutions in Super M Gag, E25K and E70K, are indicated.