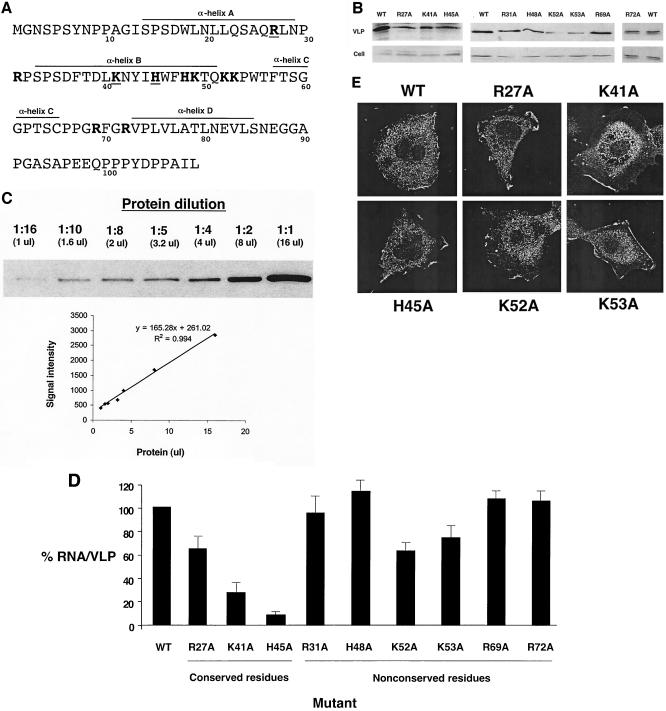
FIG. 2.
Mutagenesis of basic residues in the BLV MA domain of PrGag results in RNA packaging defects. (A) The BLV MA domain of PrGag. Amino acid residues in boldface indicate basic residues, and boldface residues that are underlined indicate basic and/or charged residues conserved among BLV, HTLV-1, and HTLV-2. The α-helices in MA are indicated, based upon nuclear magnetic resonance solution structure analysis of BLV MA (28). (B) Quantitation of VLP production. Preparation of lysates from VLP-producing cells and VLPs and immunoblot analysis are described in Materials and Methods. Representative results from the immunoblot analysis are shown. WT, wild type. (C) Linear range of detection for immunoblot analysis. Protein was diluted (1:1, 1:2, 1:4, 1:5, 1:8, 1:10, and 1:16) and was subjected to immunoblot analysis. The band intensity (arbitrary units) for each dilution was determined with the Quantity One software package of the Chemi Doc 2000 Documentation System (Bio-Rad) and plotted against the amount of protein used for immunoblot analysis (16, 8, 4, 3.2, 2, 1.6, or 1 μl) to determine if protein detection within the dilution range was linear. (D) Relative RNA packaging efficiencies of BLV MA mutants. RNA packaging efficiencies were determined by the ratio of viral RNA to the amount of Gag from VLPs (see Materials and Methods for details). The RNA packaging efficiency with wild-type BLV MA was set at 100, and the values for the MA mutants (± standard deviation) are relative to that for the wild type. Mutants were tested in parallel with the parental construct, and each experiment was done in triplicate. (E) Cellular distribution of PrGag for selected MA mutants. Cells stably transfected with VLP constructs containing mutations in MA were grown on coverslips, fixed, and incubated with an anti-HA Ig followed by incubation with Alexa Fluor 488-conjugated antimouse Ig. Images were obtained with a confocal microscope.