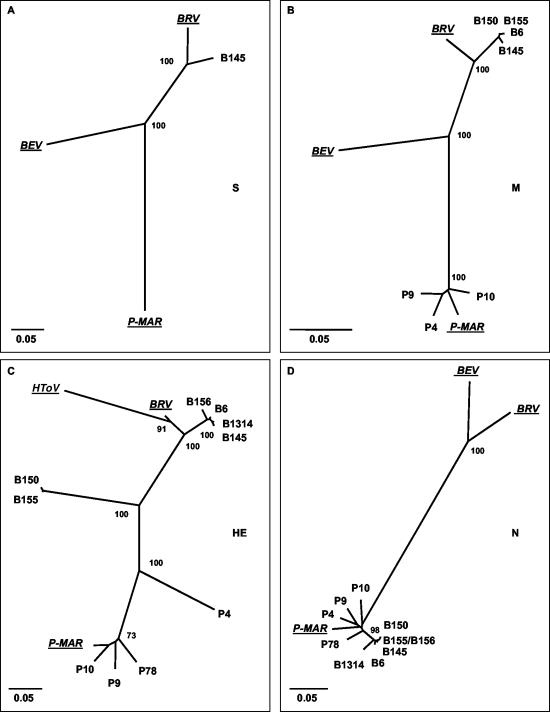
FIG. 3.
Unrooted neighbor-joining trees depicting the phylogenetic relationships among torovirus field variants. Trees were constructed for (A) the S gene, (B) the M gene, (C) the HE gene, and (D) the N gene with the Kimura-2 parameter method. Confidence values calculated by bootstrap analysis (1,000 replicates) are indicated at the major branching points. Branch lengths are drawn to scale; the scale bar represents 0.05 nucleotide substitution per site. The torovirus reference strains BRV, BEV, P-MAR, and HToV are italicized and underlined. The tree shown for the HE gene was based upon an alignment corresponding to residues 1 to 1049 of B145. Note that BEV was not included in this tree; as a result of a large deletion, Berne virus has lost most of its HE gene (51).