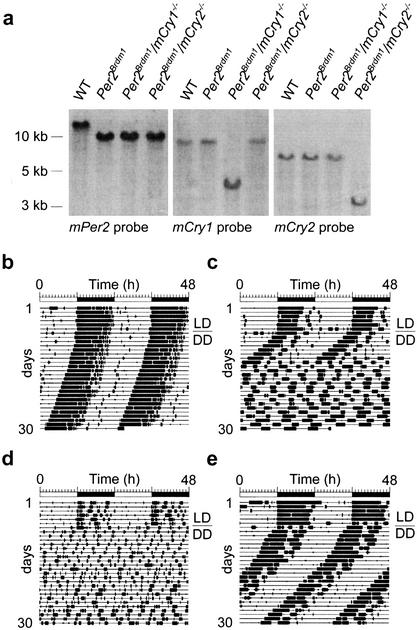
Figure 1.
Generation of mPer/mCry double mutant mice and representative locomotor activity records. (a) Southern blot analysis of wild-type (WT), Per2Brdm1, Per2Brdm1/mCry1−/−, and Per2Brdm1/mCry2−/− tail DNA. The Per2 probe hybridizes to a 12-kb wild-type and a 10-kb mutant fragment of BamHI-digested genomic DNA. The mCry1 probe detects a 9-kb wild-type and a 4-kb NcoI-digested fragment of the targeted locus. In mCry2 mutants, the wild-type allele is detected by hybridization of the probe to a 7-kb EcoRI fragment, whereas the mutant allele yields a 3.5-kb fragment. The left panel indicates size of DNA fragments. (b) Representative locomotor activity records of wild-type (WT), Per2Brdm1, Per2Brdm1/mCry1−/−, and Per2Brdm1/mCry2−/− mice. All animals were kept in a 12-h light:12-h dark cycle (LD) for at least 7 d before release into constant darkness (DD, indicated by the line over the DD). Activity is represented by black bars (three plot heights, >1, >10, and >20 wheel revolutions per 5-min period) and is double-plotted. The top bar indicates light and dark phases in LD. For the first 5 d in DD, wheel rotations per day were 20,021 ± 2,524 (n = 22) for wild-type animals, 17,656 ± 3,301 (n = 17) for Per2Brdm1 mutants, 16,025 ± 3,201 (n = 12) for Per2Brdm1/mCry1−/− mutants, and 17,859 ± 2,703 (n = 15) for Per2Brdm1/mCry2−/− mutants.