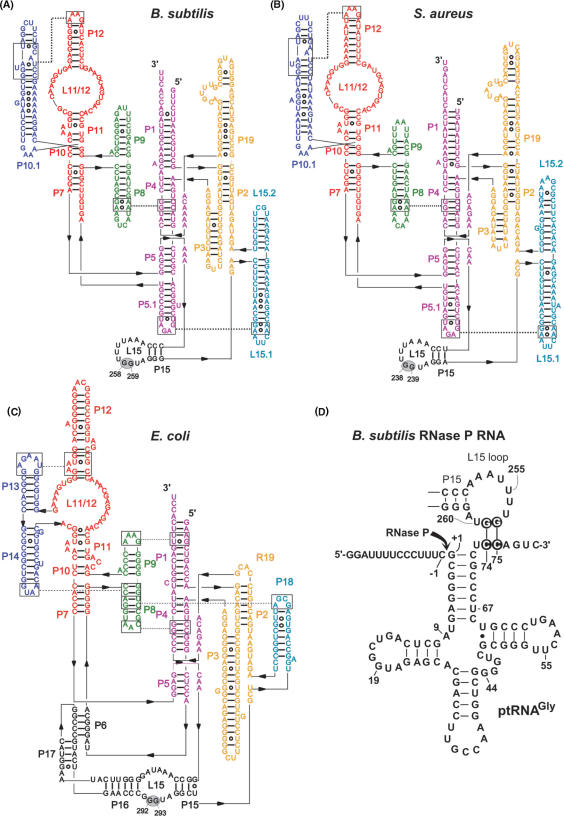
Figure 1.
Secondary structure illustrations of (A) Bacillus subtilis (type B), (B) Staphylococcus aureus (type B) and (C) Escherichia coli (type A) RNase P RNA according to (7). The two G residues in L15, known (E. coli) or suspected (B. subtilis, S. aureus) to be involved in the interaction with tRNA 3′-CCA, are highlighted by gray ovals. (D) Proposed interaction of a canonical bacterial ptRNA (Thermus thermophilus ptRNAGly) with the L15 loop of B. subtilis RNase P RNA. Highlighted nucleotides mark the sites of mutation investigated in this study. The arrow indicates the canonical RNase P cleavage site (between nucleotide –1 and +1).